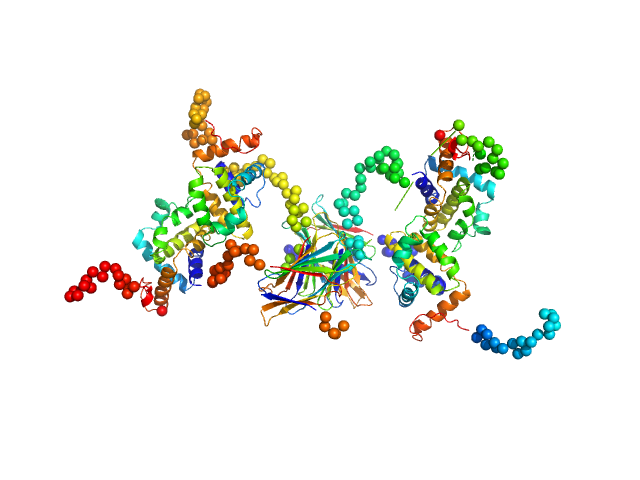
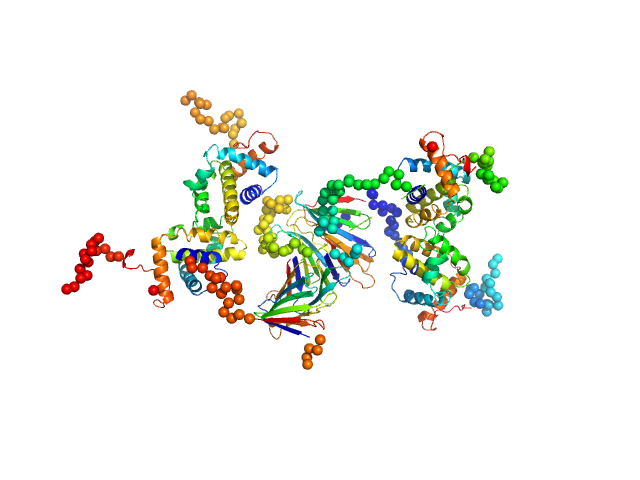
|
Synchrotron SAXS data, I(s) vs s (s = 4π sin θ/λ, where 2θ is the scattering angle) were collected from a sample of full-length Vaccinia virus A46 protein using continuous-flow size-exclusion chromatography SAXS (SEC-SAXS; Superdex 200 10/300 column) at the BM29 beam line on the ESRF storage ring (Grenoble, France). Data were collected using a Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.09918 nm. The SEC mobile phase consisted of 20 mM Tris-HCl, 10 mM DTT, pH 8.5, (20°C) with a sample injection concentration of 4.4 mg/ml. Data obtained from solute-free eluates and SEC-elution peak were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted from the SEC-peak frames to produce the data displayed in this entry.
Storage temperature = UNKNOWN. Number of frames = UNKNOWN
|
|
Protein A46
(VACV A46)
|
| Mol. type |
|
Protein |
| Organism |
|
Vaccinia virus |
| Olig. state |
|
Tetramer |
| Mon. MW |
|
28.0 kDa |
| |
| UniProt |
|
P26672
|
| Sequence |
|
FASTA |
| |
|
PDB ID
|
|
4LQK
|
| |
|
PDB ID
|
|
4LQK
|
| |
|
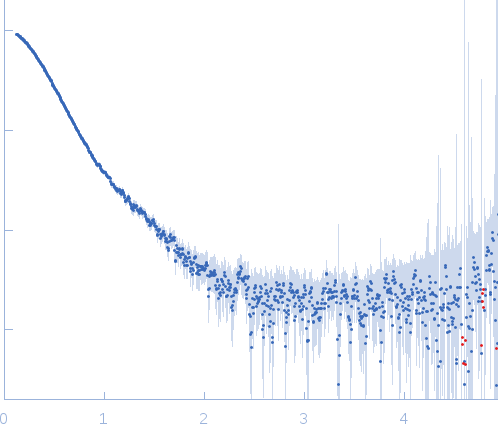 s, nm-1
s, nm-1