|
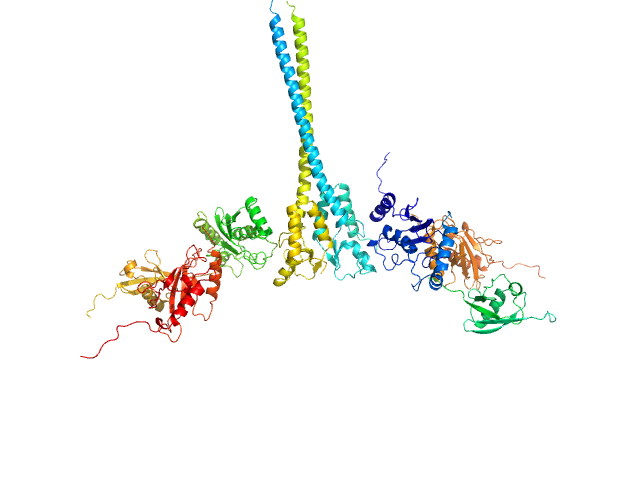
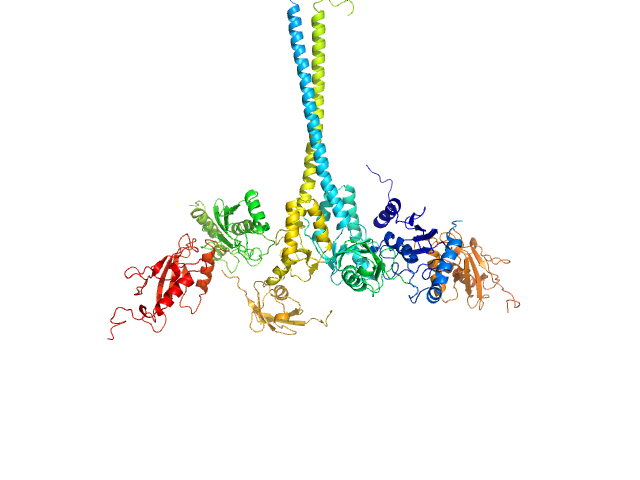
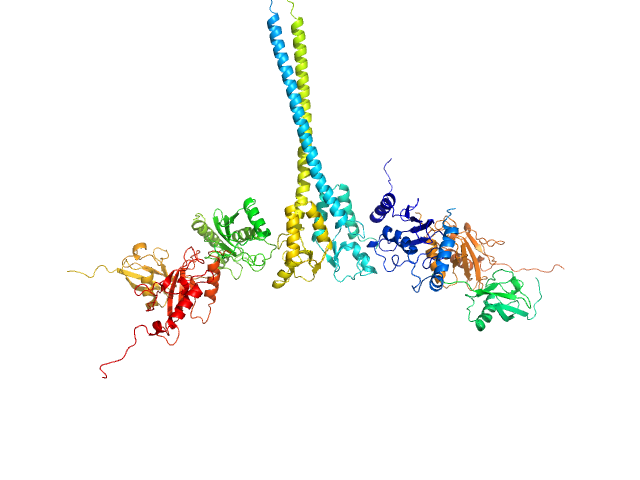
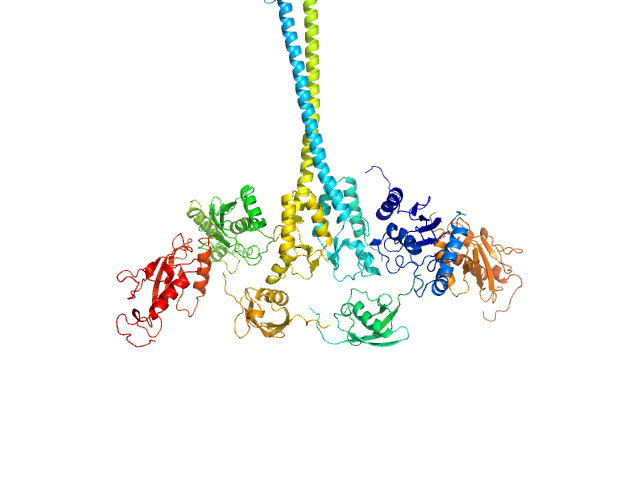
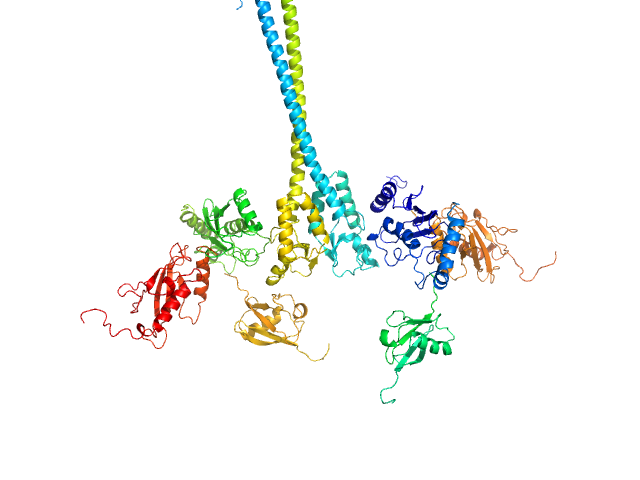
X-ray synchrotron radiation scattering data from solutions of a 2:2:2:2 complex of E3 ubiquitin-protein ligase RNF8 (L451D mutant), Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A), Ubiquitin-conjugating enzyme E2 variant 2 and Polyubiquitin-C in 20 mM HEPES 200 mM NaCl, 0.01 mM, ZnSO4, and 1 mM DTT were collected on the BL12.3.1 SIBYLS camera on the storage ring ALS (Berkeley, CA, USA) using a Pilatus 2M detector (s = 4π sin θ/λ, where 2θ is the scattering angle). The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted and the curves were scaled for protein concentration. The models shown in this entry were generated in BILBOMD from an initial pool of 7195 structures. The theoretical scattering profiles for those models were calculated using FoXS, and FoXS was also used to run a minimal ensemble search (MES). The percent contribution to the MES is written in the header of each of the PDB file, expressed as a volume fraction. For the models displayed in this entry, the volume-fractions are (from top to bottom): 0.29, 0.28, 0.21, 0.20 and 0.02. The fit of the MES was calculated using FoXS.
Cell temperature = UNKNOWN. Storage temperature = UNKNOWN. Sample detector distance = UNKNOWN. X-ray Exposure time = UNKNOWN. Number of frames = UNKNOWN. Concentration min = UNKNOWN. Concentration max = UNKNOWN
|
|
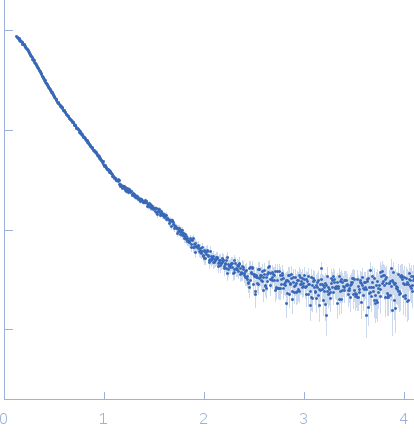 s, nm-1
s, nm-1