|
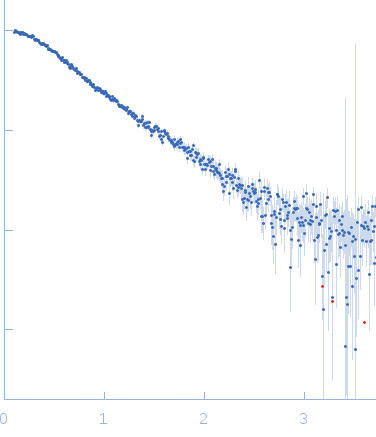
SAXS data were acquired from solutions of duplex methylated C-terminal ZBTB38 binding sequence (mCZ38BS) in 10 mM Tris (pH 6.8), 1 mM tris(2-carboxy-ethyl)phosphine (TCEP), 0.05% NaN3 and 10% D2O. The following complimentary DNA oligonucleotides containing the mCZ38BS consensus site were synthesized with numbering and methylation sites as follows:1-27 5'GCACTCATmCGGmCGCAGATCAGCTAGCC-3' and 28-54 5'-GGCTAGCTGATCTGmCGCmCGATGAGTGC-3'. Duplex DNA was formed by annealing stoichiometric amounts of sense and antisense oligonucleotides. An in-house Anton Paar SAXSess equipped with a Dectris mythen microstrip detector and set with a 10 mm line focus was utilized for data acquisition. The data were desmeared by multiplication point by point by the ratio of the desmeared to smeared model P(r) profiles calculated using GNOM with a parameterised slit correction (AH = 2.9 nm-1, LH = 1.5 nm-1) and checked for accuracy against the Anton Paar SAXSQuant desmearing routine based on the Lake algorithm. Scattering data are on an absolute scale (cm-1) set by reference to the scattering from pure H2O. The concentration of the mCZ38BS (determined by UV absorption at 260 nm) was 4.2 mg/mL. Molecular mass estimates from I(0) and Porod volume were within 5-10% of the values expected based on sequence. MULCh was used to calculate the average partial specific volume (0.591 mL/g) and X-ray contrast (Δρ = 5.4e10 cm-2). DAMMIN calculations showed an extended shape that is consistent with the expected duplex DNA structure
Storage temperature = UNKNOWN
|
|
 s, nm-1
s, nm-1
