| MWexperimental | 50 | kDa |
| MWexpected | 55 | kDa |
| VPorod | 83 | nm3 |
|
log I(s)
1.72×102
1.72×101
1.72×100
1.72×10-1
|
 s, nm-1
s, nm-1
|
|
|
|

|
|
|
|
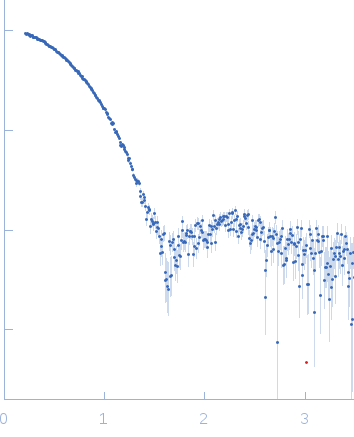
Synchrotron SAXS
data from solutions of
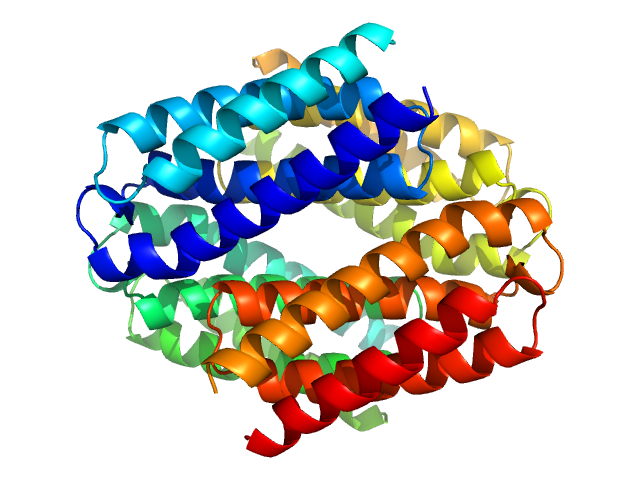
Nmul_A1745 protein from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR72
in
5 mM DTT 100 mM NaCl 10 mM Tris-HCl 0.02 % NaN3, pH 7.5
were collected
on the
BL4-2 beam line
at the Stanford Synchrotron Radiation Lightsource (SSRL) storage ring
(Menlo Park, CA, USA)
using a Rayonix MX225-HE detector
at a sample-detector distance of 1.5 m and
at a wavelength of λ = 0.13 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 0.7 and 2.2 mg/ml were measured
at 20°C.
20 successive
1 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
|
|
|||||||||||||||||||||||||||||||||