|
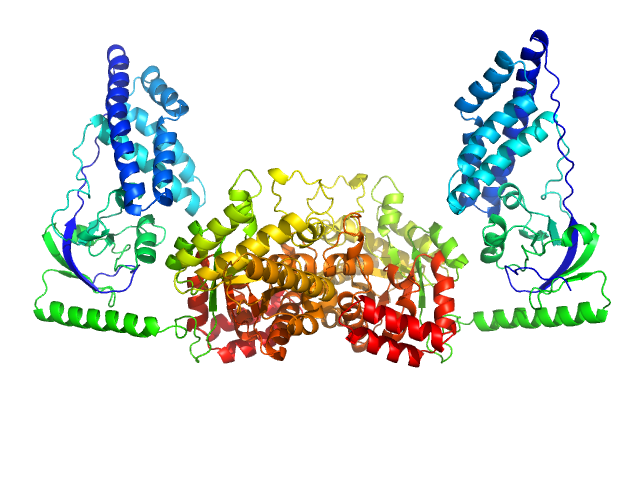
Synchrotron SAXS data from a solution of Phosphoenolpyruvate-protein phosphotransferase in 20 mM TRIS buffer, pH 7.4, 100 mM NaCl, 10 mM DTT, 4 mM MgCl2, 1 mM EDTA, and 1 tablet of protease inhibitor cocktail (SigmaFAST S8830) were acquired at the beamline 12ID-C at the Advanced Photon Source (APS, Argonne, IL, USA). One solute concentration of 5 mg/ml (corresponding to ∼40 μM dimer) was measured at 25°C using a Gold CCD detector at a sample-detector distance of 4 m and at a wavelength of λ = 0.06199 nm (s = 4π sin θ/λ, where 2θ is the scattering angle). 20 successive 0.25 second frames were collected. To prevent radiation damage, volumes of 150 μL of samples and buffers were oscillated during data collection using a flow-though setup. Individual data frames were masked, corrected for the detector sensitivity, radially integrated and normalized by the corresponding incident beam intensities. The final 1D scattering profiles and their uncertainties were calculated as means and standard deviations over the 20 individual frames. The buffer data were then subtracted from the scattering profiles.
Storage temperature = UNKNOWN
|
|
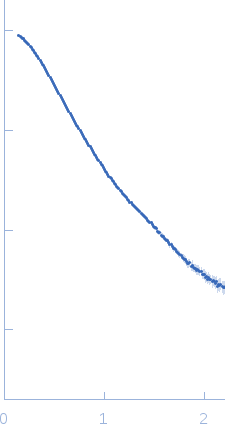 s, nm-1
s, nm-1