|
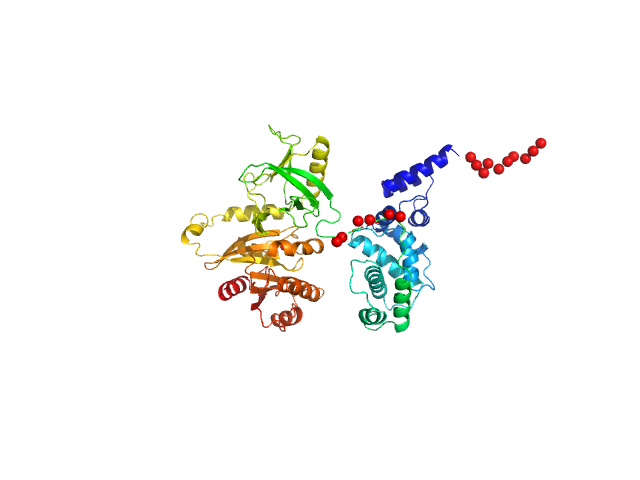
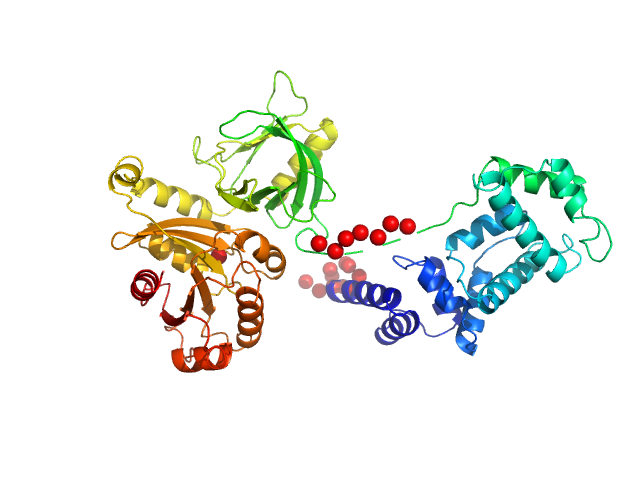
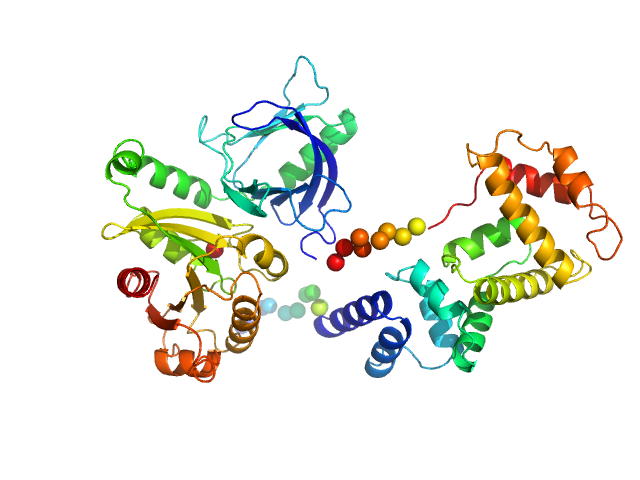
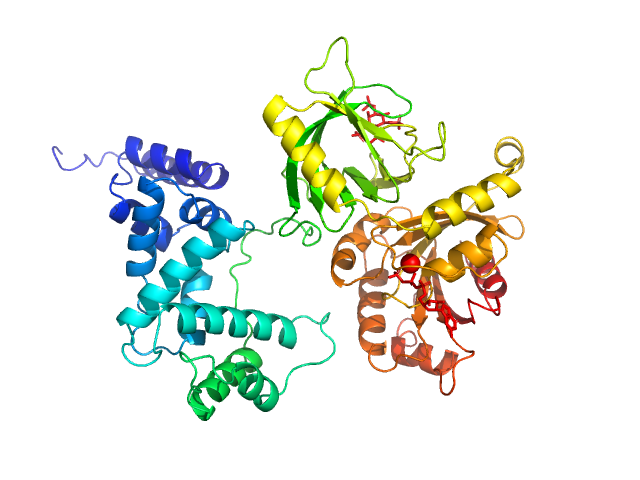
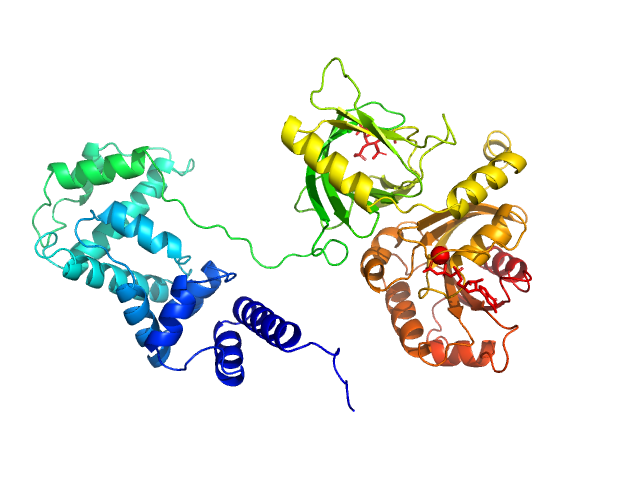
This experimental profile was derived from SEC-SAXS data collected at the BioCAT Sector 18-ID beamline at the Argonne National Laboratory Advanced Photon Source using a MAR 165 CCD detector. The protein was equilibrated with a 1.2 molar excess of inositol 1,3,4,5-tetrakis phosphate (IP4), concentrated to 10 mg/ml, and injected in 0.1 ml onto a 3 ml Superdex-200 Increase column equilibrated with 20 mM Tris, pH 8.0, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, and 0.001 ml IP4. SAXS data sets were acquired with 1 s exposures at 5 s intervals during elution at a flow rate of 0.25 ml/min. Raw SAXS images were radially averaged on a log scale over the q range 0.00621-0.333 Å-1 and normalized by the incident beam intensity. The protein scattering profile was reconstructed by singular value decomposition and linear combination (SVD-LC) as described in Malaby et al. (2015) Methods for analysis of size-exclusion chromatography-small angle X-ray scattering and reconstruction of protein scattering. J Appl Crystallogr 48: 1102-1113.
Storage temperature = UNKNOWN. Concentration min = UNKNOWN
|
|
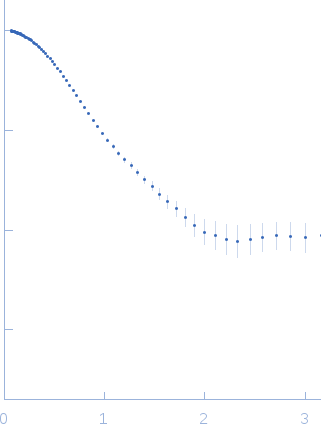 s, nm-1
s, nm-1
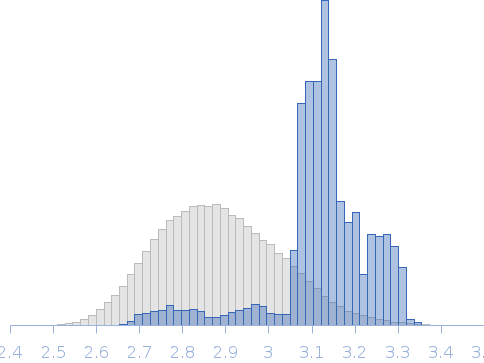 Rg, nm
Rg, nm