|
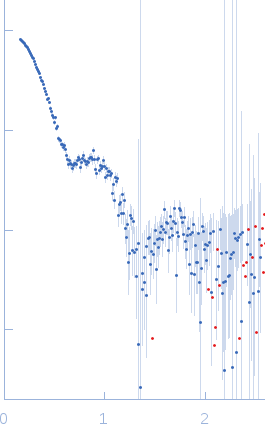
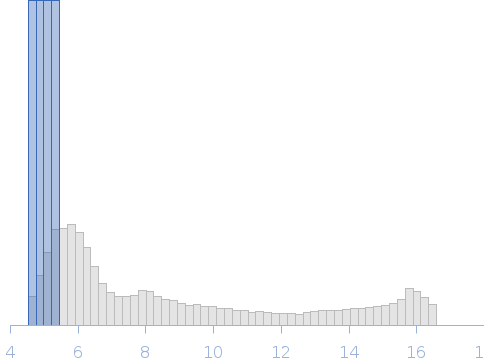
Synchrotron SAXS data from solutions of 12N12 nucleosome in 60% sucrose with ADP-BeF3 in 10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH 7.8, were collected on the G1 beam line at the Cornell High Energy Synchrotron Source (Ithaca, NY, USA) using a Pilatus Pilatus 200K detector at a sample-detector distance of 2.1 m and at a wavelength of λ = 0.1109 nm (I(s) vs s, where s = 4πsinθ/λ and 2θ is the scattering angle). One solute concentration of 1.10 mg/ml was measured at 25°C. 40 successive 10 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted to produce the scattering profile displayed in this entry.
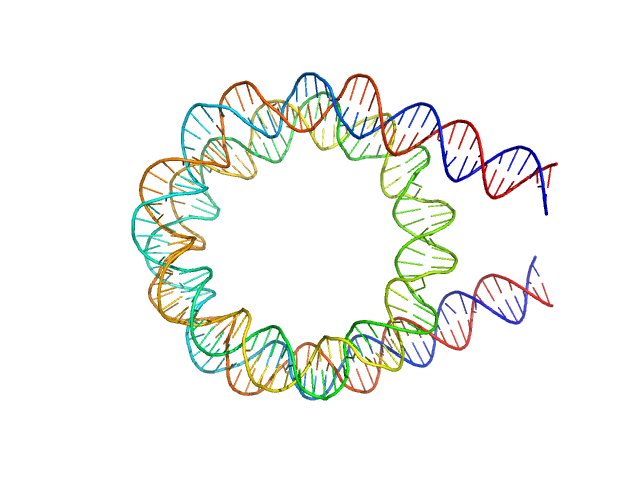
12N12 nucleosome measured in 60% sucrose and ADP-BeF3.
Due to contrast matching between the solvent and histone proteins (due to sucrose), the measured signal is dominated by the DNA component. The model was generated using 3D-DART and custom scripts written in Pymol. The percentage-weighting of each model in the ensemble ('XXXperc') is noted in the model key below:
SASDCT6_fit1_model1 = 12N12_ADP_01_100perc
|
|
 s, nm-1
s, nm-1
 Rg, nm
Rg, nm