|
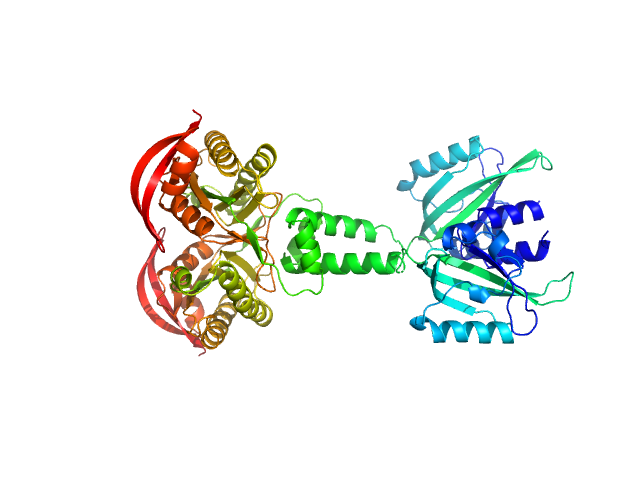
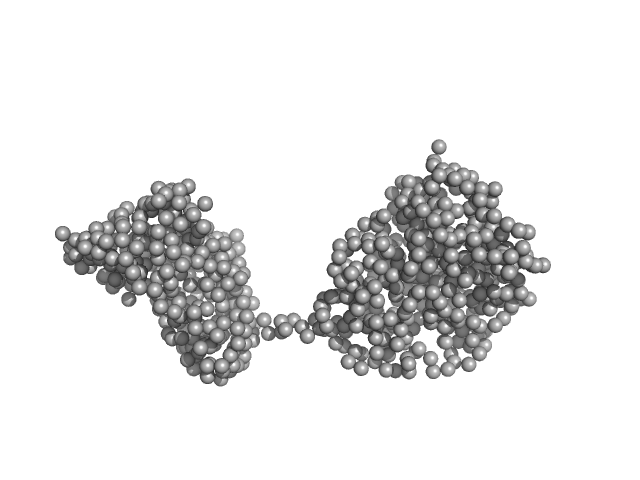
Synchrotron SAXS measurements (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle; λ = 0.11 nm) were carried out at the beam line X12SA of the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland). Quartz capillaries (inner diameter 1 mm) were mounted in a sample holder and filled with buffer (20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 5 % w/v glycerol, 1 mM ApCpp, pH 7.5). The sample holder was cooled to 10 °C for all measurements. Scattering of the buffer was determined at 20 positions along the capillary (spacing 0.5 mm), exposing each position with 11.2 keV photons for 0.5 s using 200 successive frames. Each position was measured 10 times and scattering intensities, after radial averaging to produce 1D-scattering patterns, were averaged. Scattered X-rays were detected by a Pilatus 2M detector (Dectris, Baden-Dättwil, Switzerland) at the end of an evacuated flight tube, 2.13 m from the sample position. Buffer was replaced by the protein sample without moving the sample holder or the capillaries. Lights in the experimental hutch were switched off and after 5 minutes, scattering of the protein-containing solutions was measured at the same positions using the procedure described above. Solute concentrations ranging between 5.1 and 9.1 mg/ml were measured and the concentration series data extrapolated to infinite dilution generating the final SAXS profile displayed in this entry. GASBOR was run 10 times to generate bead-model reconstructions (an averaged representation, top, and individual model, bottom).
Storage temperature = UNKNOWN
|
|
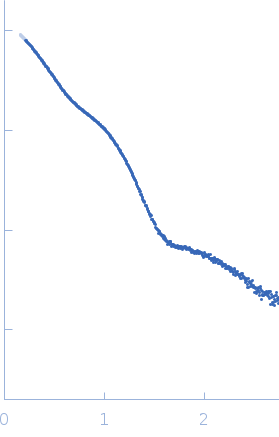 s, nm-1
s, nm-1