|
Synchrotron SAXS data from a solution of the Bc-TubR:S48 DNA complex in 0.1 M NaCl, 10 mM Tris, pH 8 were collected on the BL-10C beam line at the Photon Factory (PF), High Energy Accelerator Research Organization (KEK) storage ring (Tsukuba, Japan) using a Pilatus3 2M detector at a sample-detector distance of 2.0 m and at a wavelength of λ = 0.15 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Data were collected for a 20 s X-ray exposure time at a sample temperature of 8°C. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
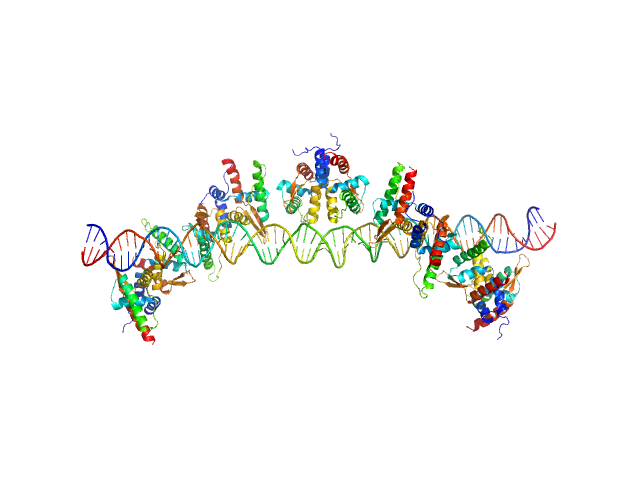
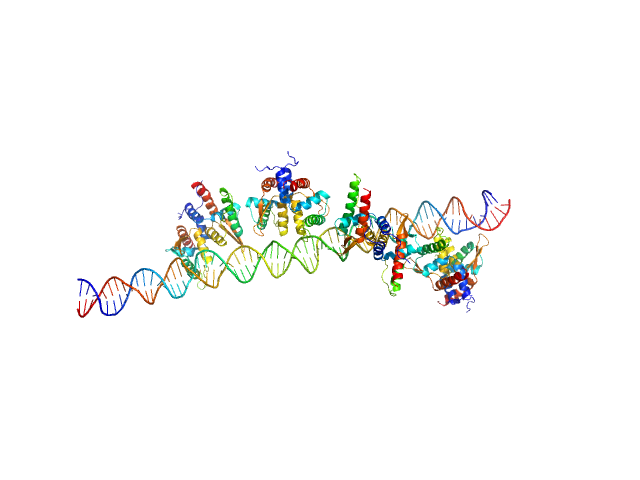
The molecular weight (MW) of the Bc-TubR : S48 DNA complex was estimated from the macromolecular volume using the method of Fischer et al. (J. Appl. Crystallogr. 43, 101–109, 2010; SAXMow program). However, the MW estimate is likely not an accurate value because the sample is a complex of protein and DNA. The theoretical SAXS curve of the molecular dynamics model was calculated as the mixture of the two models (one model contains four TubR dimers plus DNA, another model contains five TubR dimers plus DNA). For the mixed model, the fractions of each are 0.4 and 0.6, respectively. A low resolution dummy atom model was built by DAMMIF. Ten independently built models were generated, spatially aligned and then bead-occupancy and volume corrected using DAMAVER to generate an averaged spatial representation of the complex (damfilt). The the fit to the SAXS data from an individual dummy atom model from the cohort of models used to obtain the average representation is shown, specifically that with the smallest chi-square value.
Sample concentration: UNKNOWN
|
|
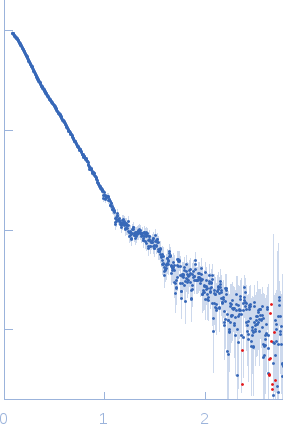 s, nm-1
s, nm-1