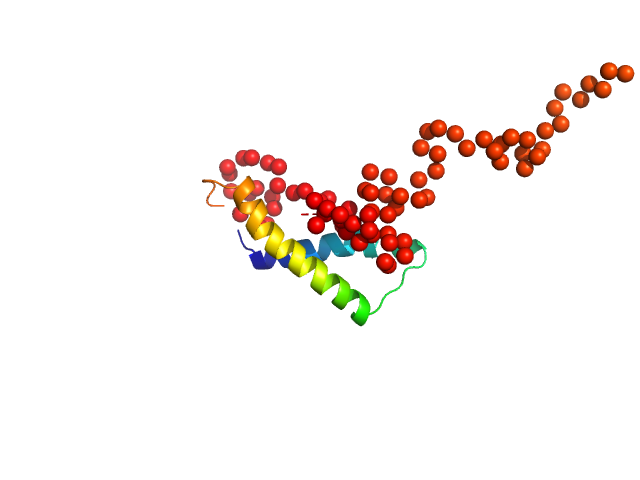|
Synchrotron SAXS data from solutions of the C-terminal truncated bank vole prion protein (amino acids 90-231) in 25 mM ammonium acetate, 250 mM NaCl, pH 5.5 were collected using size exclusion chromatography SAXS (SEC-SAXS) on the BM29 beam line at the ESRF (Grenoble, France) using a Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.099 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). SEC-SAXS was performed at 20 °C using the following parameters: Column type, Superdex 75 10/300 GL (GE Healthcare); Flow rate: 0.5 mL/min; Total acquisition time: 50 min; Injection concentration: 10 mg/mL; Injection volume: 250 μL. The data obtained through the sample elution peak (1963-2030 1 s frames) were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted and the individual subtracted data sets were scaled and averaged to generate the scattering profile displayed in this entry.
SEC column = UNKNOWN. Sample injection volume = UNKNOWN. Flow rate = UNKNOWN
|
|
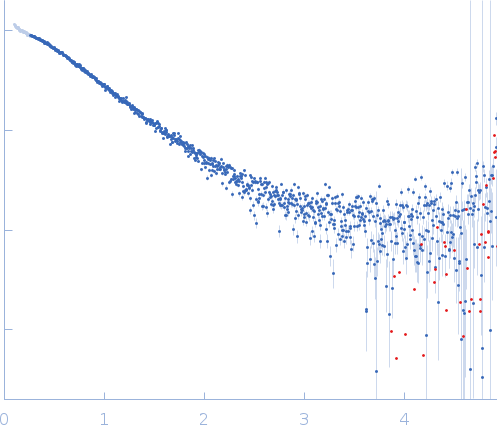 s, nm-1
s, nm-1
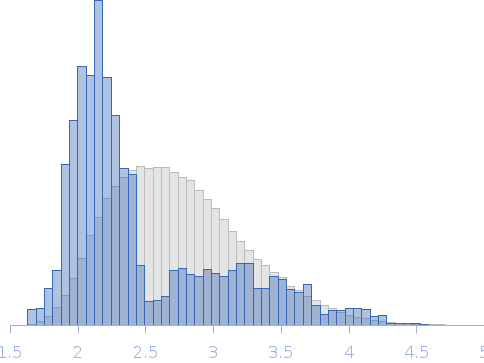 Rg, nm
Rg, nm