|
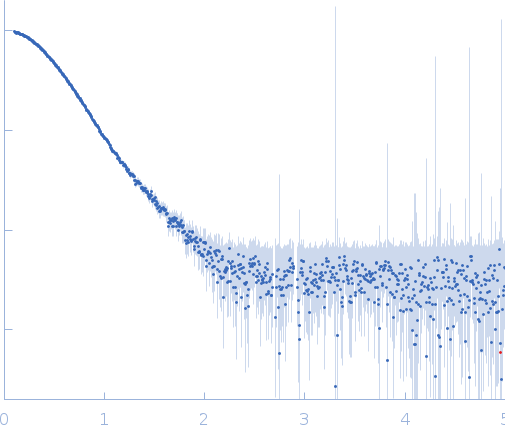
Synchrotron SAXS data from solutions of GON7 bound to LAGE3-OSGEP in 20 mM MES, 200 mM NaCl, 5 mM β-mercaptoethanol, pH 6.5 were collected on the SWING beam line at SOLEIL (Saint-Aubin, France) using a AVIEX PCCD170170 detector at a sample-detector distance of 1.8 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 65.00 μl sample at 1.5 mg/ml was injected at a 0.30 ml/min flow rate onto a Agilent Bio SEC-3, 300 Å column at 15°C. 255 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The scattered intensities were displayed on an absolute scale (cm-1) using the scattering of water. SAXS data were normalized to the intensity of the incident beam, averaged and background subtracted using the program US-SOMO. Frames were examined individually and 21 identical frames were averaged and further processed. The corresponding SEC-elution concentration was approximately 0.35 mg/mL.
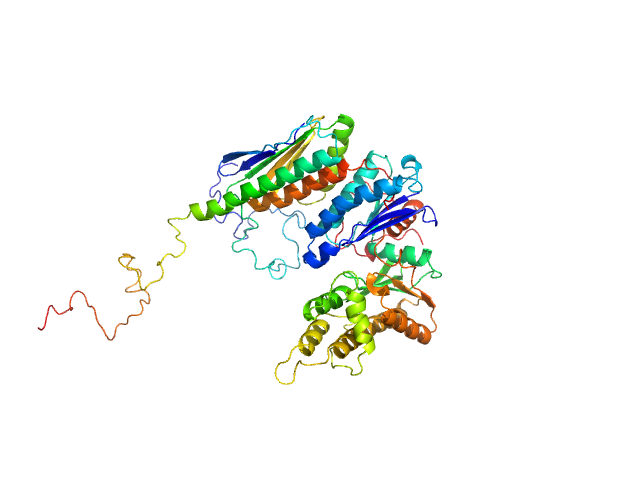
The complex is constituted of one monomer of OSGEP, one monomer of LAGE3 and one monomer of GON7. Molecular mass was obtained using the Bayesian inference approach proposed in PRIMUS/qt, which combines four concentration independent MM estimators: M=59.5 kDa within the credibility interval [57.5-64.5].
|
|
 s, nm-1
s, nm-1