|
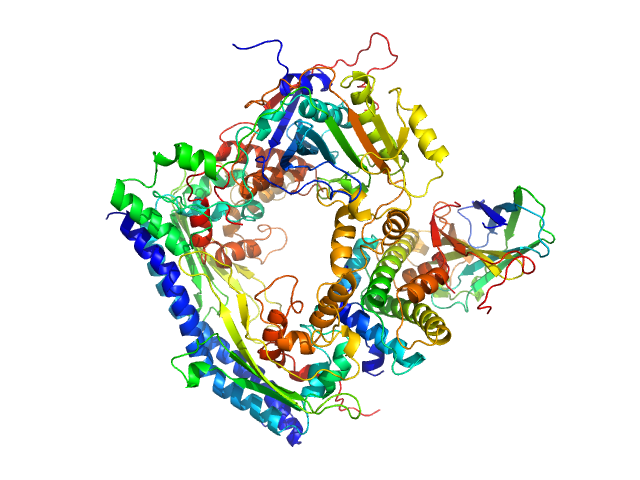
SANS data from solutions of the Asf1-H3:H4-Rtt109-Vps75 protein complex (protonated Asf1-H3:H4 bound to perdeuterated Rtt109-Vps75) in 50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pD 6.5 were collected on the KWS1 instrument at FRM2 (Munich, Germany) using a SANS 6Li-Scintillator 1 mm thickness + photomultiplier detector. Due to a combination of negative (from 1H Asf1-H3:H4) and positive (from perdeuterated Rtt109-Vps75) contrasts the Rg obtained from the Guinier approximation is negative. The data were recorded at two detector configurations: 1) 4 m for 6 hrs using a neutron wavelength of 0.5 nm and; 2) 1.5 m for 2.5 hrs using a neutron wavelength of 0.5 nm. Both measurement were performed on the same sample at 3.85 mg/mL, measured at 25°C.
Number of frames = UNKNOWN
|
|
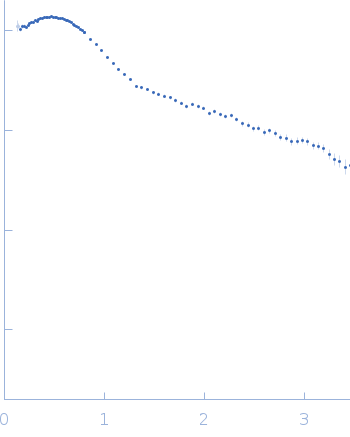 s, nm-1
s, nm-1