|
Synchrotron SAXS data from solutions of the His-RuvBl1/RuvBl2 dodecamer in 20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH 7.5 were collected using size-exclusion chromatography SAXS (SEC-SAXS) on the SWING beam line at SOLEIL (Saint-Aubin, France) using a Eiger 4M detector at a sample-detector distance of 2 m and at a wavelength of λ = 0.1033 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). SEC-SAXS was performed at 15°C using the following parameters: Column: Agilent Bio SEC-5, 500Å (4.6 mm id * 300 mm); Flow rate: 0.3 mL/min; Sample injection concentration: 8.45 mg/mL; Injection volume: 50 μL. The data were collected through the SEC peak of the protein as a series of 17 x 1 second exposures. Each unsubtracted data frame was normalised to the intensity of the transmitted beam and radially averaged and the scattering of an appropriate solvent-blank was subtracted. The resulting subtracted frames were scaled and averaged to generate the final SAXS profile displayed in this entry. The experimental molecular weight was determined from the volume of correlation, Vc.
Storage temperature = UNKNOWN
|
|
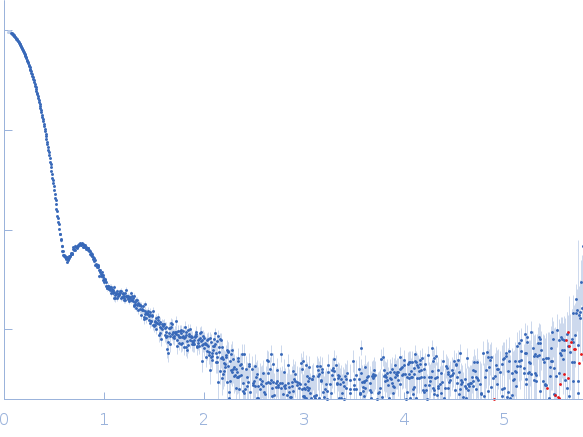 s, nm-1
s, nm-1
