|
Synchrotron SAXS data from solutions of DNA repair protein Rad5 in 40 mM Tris, 150 mM KCl, 5 mM DTT, 5% glycerol, pH 8 were collected on the BioCAT 18ID beam line at the Advanced Photon Source (APS) Argonne National Laboratory (Lemont, IL, USA) using a Pilatus3 X 1M detector at a sample-detector distance of 3.5 m and at a wavelength of λ = 0.1033 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 175.00 μl sample at 7.8 mg/ml was injected at a 0.70 ml/min flow rate onto a GE Superdex 200 Increase 10/300 column at 20°C. 19 successive 0.500 second frames were collected through the SEC elution peak of the sample. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The simulation code uiowa_BD was used to carry out Langevin dynamics (LD) simulations of Rad5. Two different sets of simulations were carried out in triplicate, with each set using a different starting model of Rad5. The different starting models were generated based on docking results from two different docking servers, ClusPro and ZDOCK. Docking was used to position the HIRAN domain on the helicase domain in an acceptable orientation. For each simulation, a time-step of 125 fs was used, and a snapshot (PDB file) was recorded every ns of simulation time for a total of 5 µs of simulation time. As such, each simulation generated a total of 5000 individual structures. The first set of models displayed in this entry (top) are representatives from the ZDOCK LD simulation while the second set of models (bottom) are representatives derived from ClusPro. A zip file containing the three replicates of each ClusPro and ZDOCK simulation are attached to this entry and made available in the full entry zip archive (5000 PDB files per simulation; ca. 2.2 GB total).
|
|
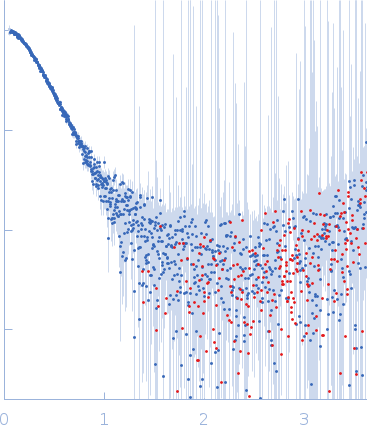 s, nm-1
s, nm-1
![Static model image DNA repair protein RAD5 OTHER [STATIC IMAGE] model](/media//pdb_file/SASDG25_fit1_model1.png)
![Static model image DNA repair protein RAD5 OTHER [STATIC IMAGE] model](/media//pdb_file/SASDG25_fit2_model1.png)