|
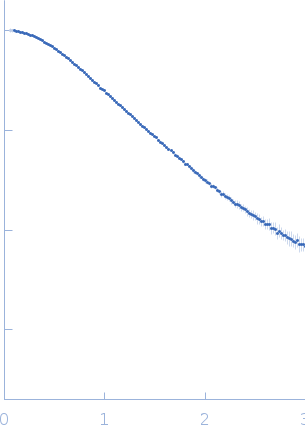
Synchrotron SAXS
data from solutions of
Xrn1 resistance RNA2 from Murray Valley Encephalitis
in
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH 7.5
were collected
on the
12-ID-B SAXS/WAXS beam line
at the Advanced Photon Source (APS), Argonne National Laboratory storage ring
(Lemont, IL, USA)
using a Pilatus 2M detector
at a sample-detector distance of 2 m and
at a wavelength of λ = 0.08857 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
Solute concentrations ranging between 0.8 and 3 mg/ml were measured
at 25°C.
30 successive
1 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The low angle data collected at lower concentrations were extrapolated to infinite dilution and merged with the higher concentration data to yield the final composite scattering curve.
The subtracted SAXS data series used to generate the final extrapolated to zero-concentration profile displayed in this entry are provided in the full entry zip archive. The ab initio dummy atom model represents the averaged spatial disposition of the RNA in solution (DAMFILT model), derived from several aligned individual models (corresponding fit).
|
|
 s, nm-1
s, nm-1
