| MWexperimental | 122 | kDa |
| MWexpected | 121 | kDa |
| VPorod | 238 | nm3 |
|
log I(s)
6.90×101
6.90×100
6.90×10-1
6.90×10-2
|
 s, nm-1
s, nm-1
|
|
|
|
 Rg, nm
Rg, nm
|
|
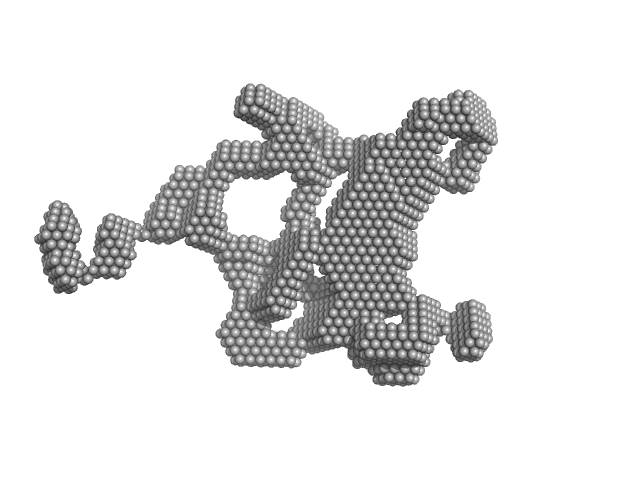
|
|
|
|
|
|
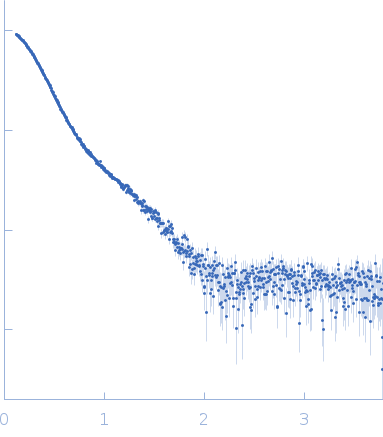
Synchrotron SAXS
data from solutions of
Nucleolar RNA helicase 2 (DDX21) fragment 186-710
in
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH 7.5
were collected
on the
BM29 beam line
at the ESRF storage ring
(Grenoble, France)
using a Pilatus 1M detector
at a sample-detector distance of 2.9 m and
at a wavelength of λ = 0.099 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 100.00 μl sample
at 10 mg/ml was injected at a 0.50 ml/min flow rate
onto a GE Superose 6 Increase 10/300 column
at 20°C.
3000 successive
1 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
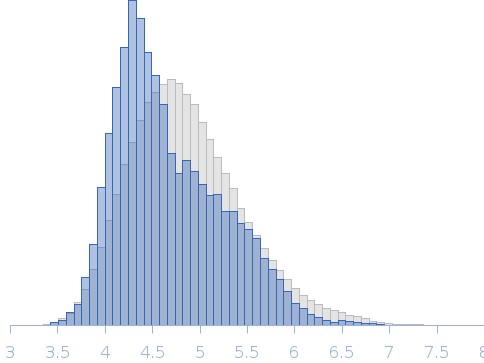
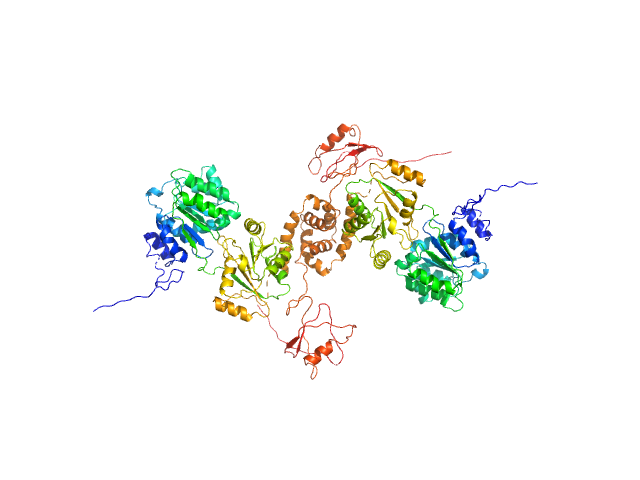
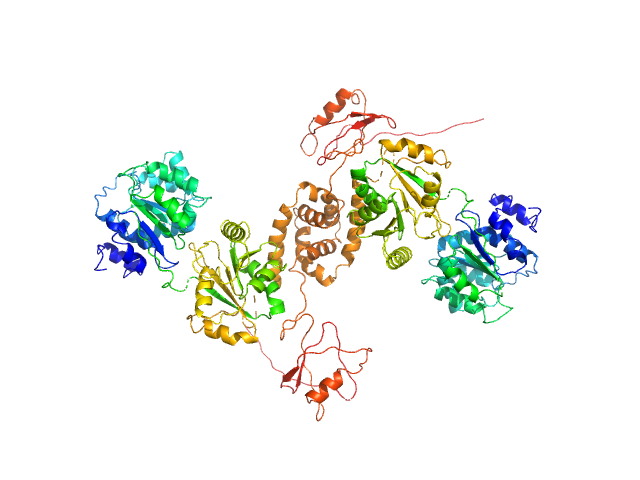
The results from ensemble optimization methos are included in the full entry zip archive. In addition, two atomistic models are shown. The first model (X^2 = 1.25) was built using two PDBs as templates in SwissModel (2M3D, 3I32). The N-terminal unstructured tags were added using I-TASSER and the model was submitted to nonlinear Cartesian Normal Mode Analysis (NOLB NMA) resulting in the final model. The second model (bottom) was created substituting the helicase domain of model 1) by the crystal structure of this domain (PDB 6l5n) and removing the His-tags. |
|
|||||||||||||||||||||||||||