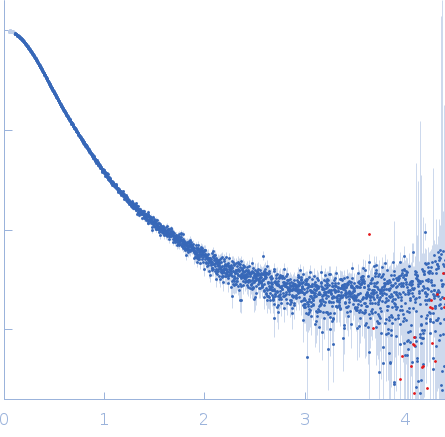
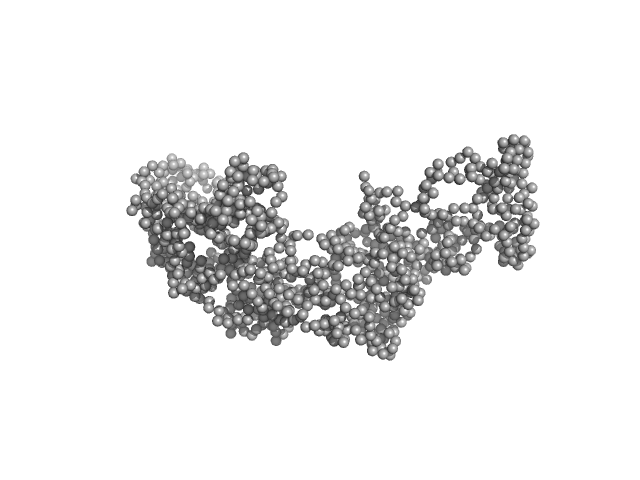
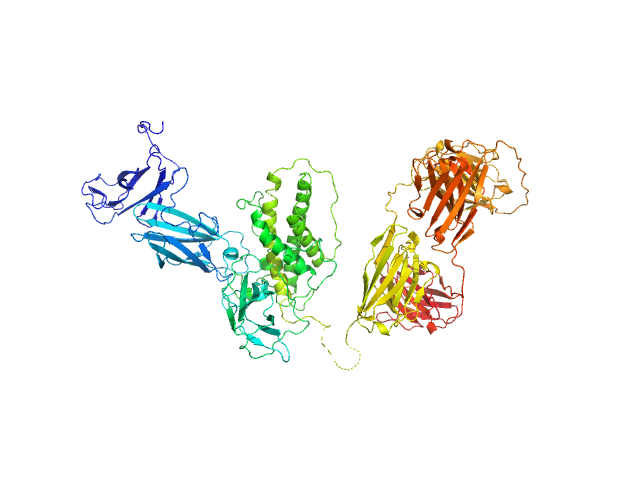
| MWexperimental | 115 | kDa |
| MWexpected | 112 | kDa |
| VPorod | 224 | nm3 |
|
log I(s)
1.20×10-1
1.20×10-2
1.20×10-3
1.20×10-4
|
 s, nm-1
s, nm-1
|
|
|
|
|
|
|
|
Synchrotron SAXS data from solutions of the IL12-L19L19 immunocytokine in 25 mM HEPES/NaOH, 0.2 M NaCl, pH 8 were collected on the B21 beam line at Diamond (Didcot, UK) at a sample-detector distance of 4.01 m and at a wavelength of λ = 0.099 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Size exclusion chromatography coupled to SAXS (SEC-SAXS) was carried out at at 20 °C using an Agilent HPLC system. 50 μL of IL12-L19L19 concentrated at 3.5 mg/mL were injected into a Superdex 200 3.2/300 PC (GE Healthcare), pre-equilibrated with 25 mM HEPES/NaOH, 200 mM NaCl, pH 8.0. 36 successive 1 second frames were collected through the sample elution peak and processed using CHROMIXS that included the identification and subtraction of an appropriate solvent-blank.
|
|
|||||||||||||||||||||