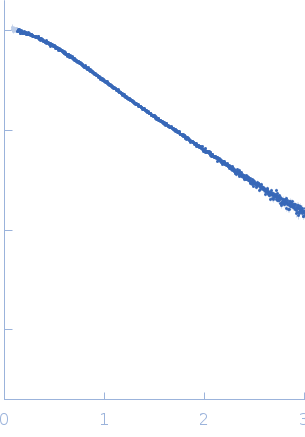
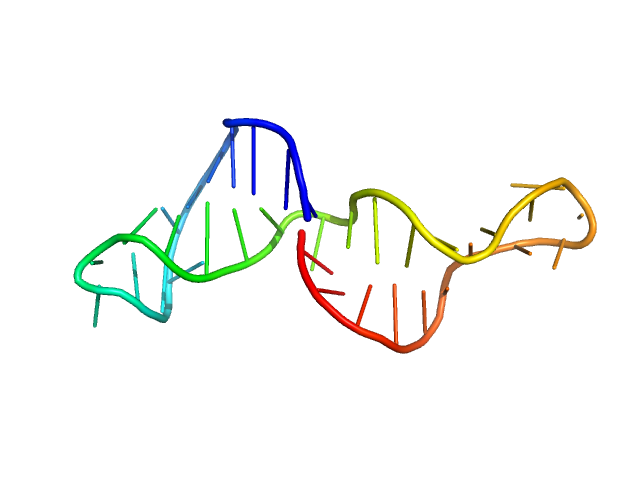
| MWexperimental | 12 | kDa |
| MWexpected | 11 | kDa |
| VPorod | 16 | nm3 |
|
log I(s)
1.24×10-2
1.24×10-3
1.24×10-4
1.24×10-5
|
 s, nm-1
s, nm-1
|
|
|
|
|
|

|
|
SAXS measurements were performed at the P12 BioSAXS beamline of the EMBL at the Petra III storage ring (DESY, Hamburg). The data were collected on a photon-counting Pilatus-6M detector at the sample-to-detector distance of 3.0 m and wavelength λ of 0.12 nm covering the momentum vector range 0.2 < s < 7.8 nm −1 (s = 4πsinθ/λ, where 2θ is the scattering angle). The scattering patterns from the dilution series were collected at four different concentrations: 8, 4, 2 and 1 mg/ml, each in a volume of 50 uL.
|
|
|||||||||||||||||||||