|
Synchrotron SAXS data from solutions of the minimal hepatocyte growth factor mimic K1K1 in 25 mM Tris, 150 mM NaCl, pH 7.4 were collected on the BM29 beam line at the ESRF (Grenoble, France) using a Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.0992 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 35.00 μl sample at 8.5 mg/ml was injected at a 0.08 ml/min flow rate onto a GE Superdex 200 Increase 3.2/300 column at 20°C. 68 successive 1 second frames were collected through the SEC-elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
SEC-SAXS experiments were carried out using Nexera High Pressure Liquid/Chromatography (HPLC; Shimadzu) system connected online to SAXS sample capillary. For these experiments, frames corresponding to protein peaks were identified, blank subtracted and averaged using CHROMIXS. Radii of gyration (Rg), molar mass estimates and distance distribution functions P(r) were computed using the ATSAS package in PRIMUS.
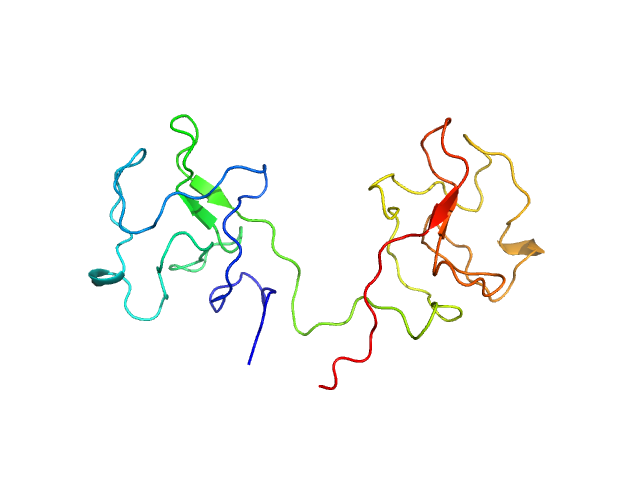
The K1K1 chimeric protein was generated by fusing two K1 domains from the N-terminal fragment of the HGF/SF (NK1) ligand.
|
|
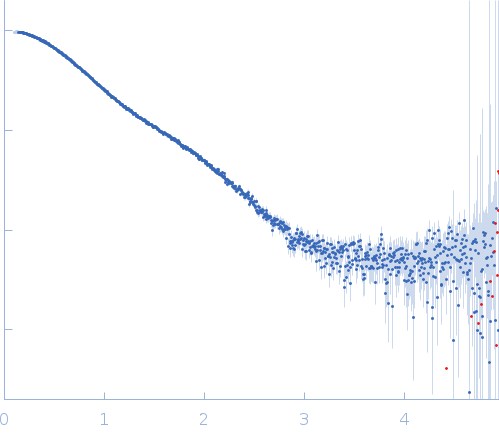 s, nm-1
s, nm-1