|
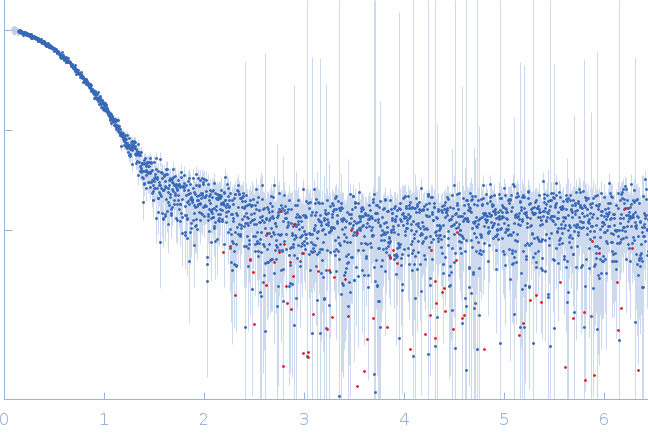
Synchrotron SAXS data from solutions of MexR in 20mM HEPES, 150mM NaCl, 10mM DTT, 1% v/v glycerol, pH 7.1 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 35.00 μl sample at 8.5 mg/ml was injected at a 0.35 ml/min flow rate onto a GE Superdex 75 Increase 5/150 column at 20°C. 63 successive 0.25 second frames were collected through the SEC elution peak of the MexR sample. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank, derived from SEC-frames measured from the elution buffer, was subtracted.
The SEC-SAXS and SEC-MALLS data are included in the full entry zip archive. The models displayed in this entry show: 1) The volume and bead-occupancy averaged spatial representation of the protein obtained from an aligned model cohort (DAMFILT). 2) An individual DAMMIF bead model of MexR and the associated fit to the data. Additional models are also provided in the full entry zip archive.
|
|
 s, nm-1
s, nm-1

