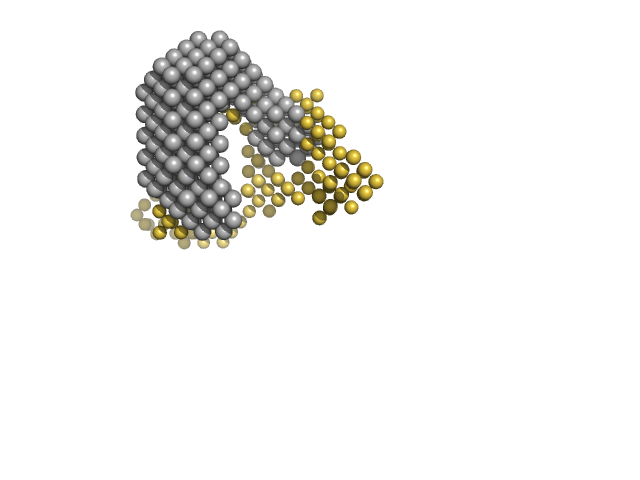|
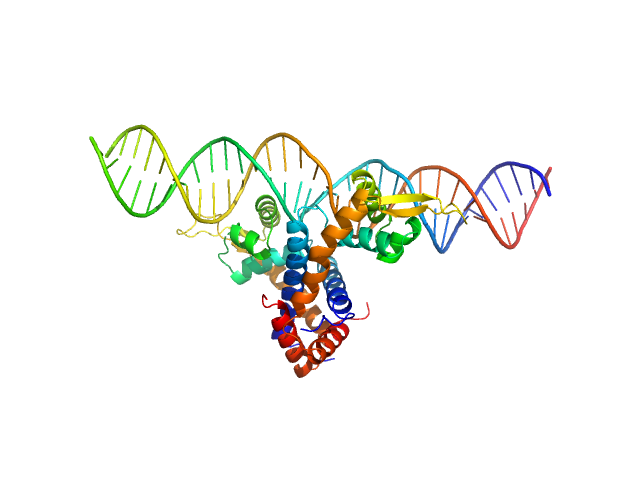
SANS data with contrast variation were collected on the D22 instrument at Institut Laue-Langevin (ILL; Grenoble, France) at the concentration of 6.2 mg/mL. In total five contrast point were measured: hydrogenated MexR (hMexR) bound to hydrogenated 34bp dsDNA (PII DNA segment) at 0, 79% v/v D2O buffer and; deuterated MexR (dMexR - 73% average non-exchangeable 2H) bound to hydrogenated 34bp dsDNA (PII DNA segment) at 0, 56, 89% D2O buffer. SAXS data of hMexR in complex with PII DNA binding region was also collected using an Anton Paar SASXess instrument (I(s) vs s, where s = 4π sin θ/λ and 2θ is the scattering angle; λ=0.154 nm) at Linköping University (Linköping, Sweden). For the SAXS measurements, approximately 40 µL of 3.12 mg/mL of protein-DNA solution was loaded into a quartz capillary and four successive 1200 second frames were collected. Matching buffer containing 20 mM sodium phosphate, 150 mM NaCl, 10 mM DTT (pH 7.1) was measured in the analogous way. Data reduction was performed with SAXS-Quant 1D software, which includes slit averaging, and normalization to absolute intensity using absolute intensity direct flux measurements, buffer-subtraction and desmearing corrections.
The SAXS/SANS data, model fits, and additional analysis are included in a downloadable zip archive. The data displayed on this entry page is of the fully hydrogenated MexR/DNA complex measured in 0% v/v D2O buffer.
|
|
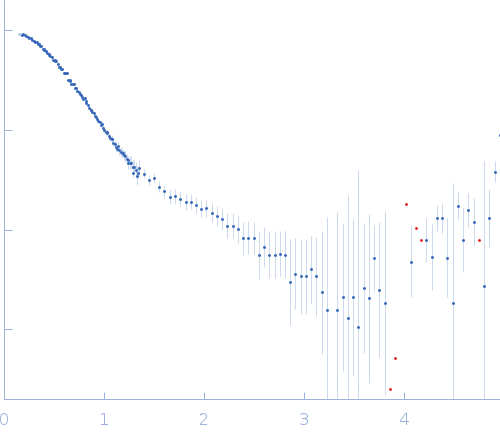 s, nm-1
s, nm-1