|
Synchrotron SAXS data from solutions of the anti-CD20 IgG monoclonal antibody (mAb) in 0.1 M HEPES, pH 7.7 were collected on the B21 beam line at the Diamond Light Source (Oxfordshire, UK) using a Pilatus 2M detector at a sample-detector distance of 4.0 m and at a wavelength of λ = 0.1 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Different solute concentration ranging from 1.00 to 6.00 mg/ml was measured at 5°C. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. Data from different concentrations were merged to get the final SAXS profile processed for modelling.
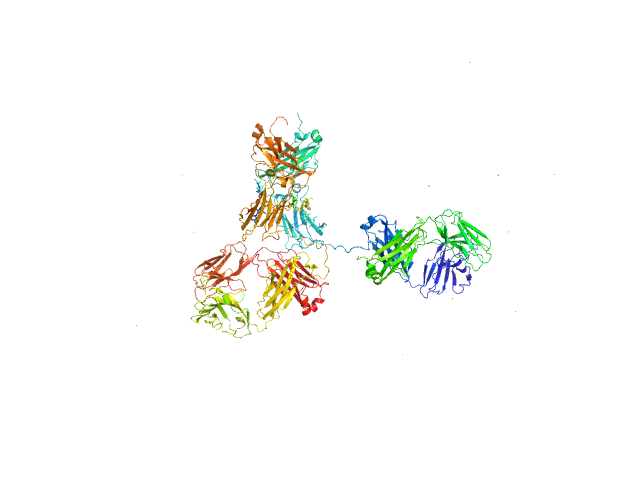
Modelling has been performed by using the averaged envelope obtained from 20 runs of DAMMIF, properly clustered, as input of a flexible fitting procedure based on molecular dynamics. The code NAMD with its extension named MDFF (molecular dynamics flexible fitting) was used. This was done to account for the extreme flexibility of the mAb, and its very peculiar shape, which is not easy to model as a molecular envelope. Ensemble analysis was performed using the ensemble optimization method (EOM). The EOM results of the resulting mAb ensemble, including selected model representatives, Rg, and Dmax distributions are located in the full entry zip archive.
This study has been carried out in the framework of the AMECRYS project (http://www.amecrys-project.eu/) - Horizon 2020 Future and Emerging Technologies programme FET-OPEN - grant agreement no.712965.
|
|
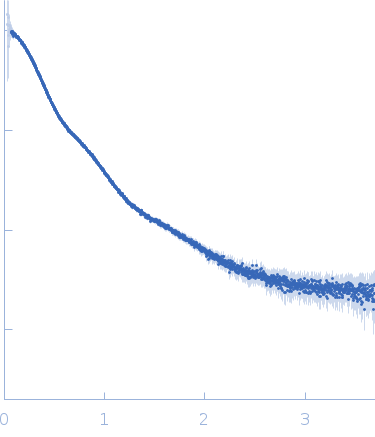 s, nm-1
s, nm-1

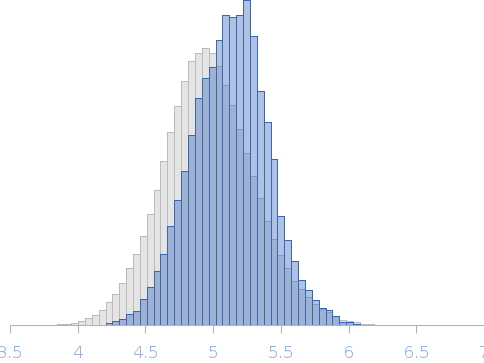 Rg, nm
Rg, nm