|
Synchrotron SAXS data from solutions of the pikachurin N-terminal FnIII(1-2) fragment in 25 mM HEPES, 200 mM NaCl, pH 8 were collected on the B21 beam line at the Diamond Light Source (Didcot, UK) using a Eiger 4M detector at a sample-detector distance of 4.0 m and at a wavelength of λ = 0.01 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 50.00 μl sample at 3 mg/ml was injected at a 0.08 ml/min flow rate onto a GE Superdex 75 Increase 3.2/300 column at 20°C. 34 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
SEC-SAXS data were analyzed using the ATSAS suite. Scattering frames corresponding to samples were selected, averaged and subtracted from averaged buffer frames using CHROMIXS. The buffer subtracted, 1D scattering curves were then processed using PRIMUS to compute the radii of gyration (Rg), and obtain molecular mass estimates; pair distribution function (P(r)) and the maximum particle dimension (Dmax) were estimated from the scattering data using the algorithm available in GNOM.
Recombinant Pika-FnIII(1-2) was produced using HEK293-F cells with an N-terminal Histidine tag. Purification was carried out using IMAC followed by gel filtration in 25 mM HEPES/NaOH, 200 mM NaCl, pH 8
|
|
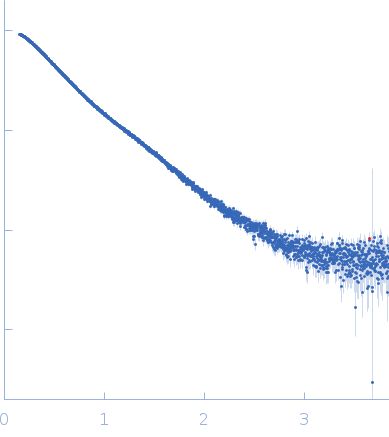 s, nm-1
s, nm-1
