|
Synchrotron SAXS data from solutions of elongation factor Tu in 20 mM Tris, 100 mM NaCl, pH 8 were collected on the BL19U2 beam line at the Shanghai Synchrotron Radiation Facility (SSRF; Shanghai, China) using a Pilatus 1M detector at a sample-detector distance of 2.6 m and at a wavelength of λ = 0.13 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 1.00 mg/ml was measured at 20°C. 20 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
The bead model shown in this entry is the volume and bead occupancy corrected model based on the alignment of an individual model cohort. It does not represent the corresponding fit to the data as displayed in this entry. CAUTION: Possibly aggregated. CAUTION: Possibly background over-subtracted.
|
|
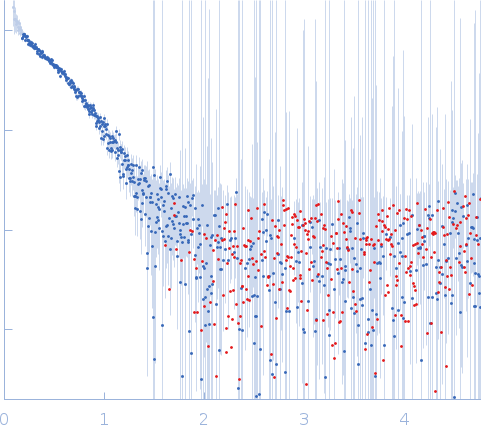 s, nm-1
s, nm-1
