| MWexperimental | 98 | kDa |
| MWexpected | 68 | kDa |
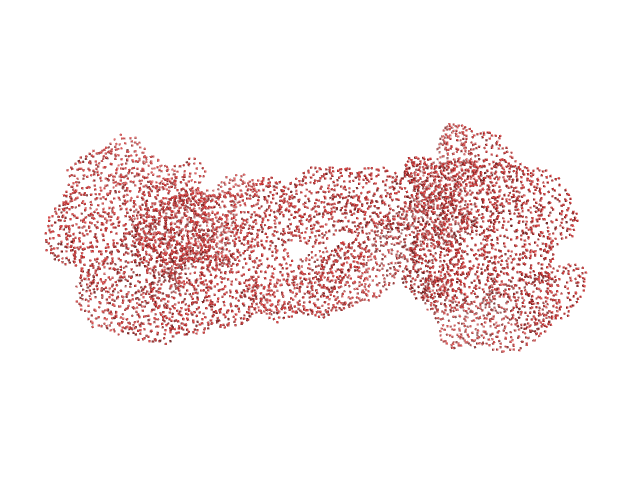
| VPorod | 230 | nm3 |
|
log I(s)
3.86×10-2
3.86×10-3
3.86×10-4
3.86×10-5
|
 s, nm-1
s, nm-1
|
|
|
|
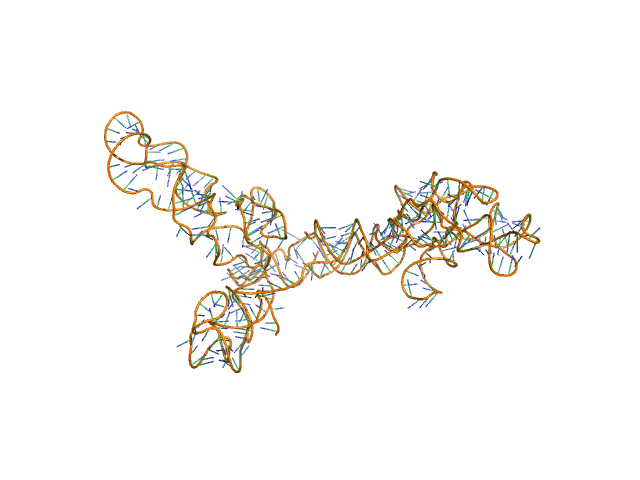
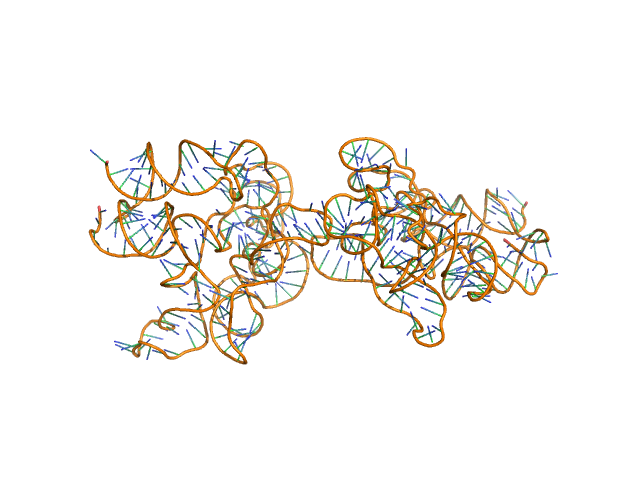
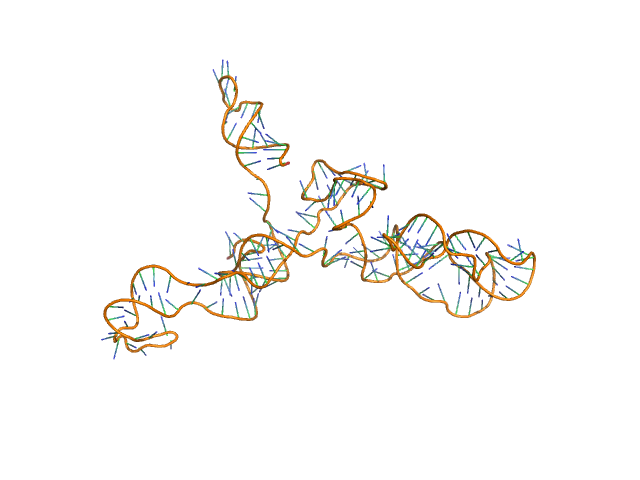
|
|
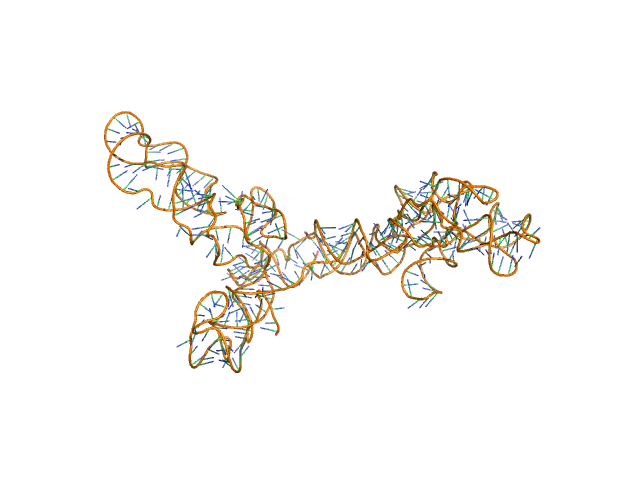
|
|
|
|
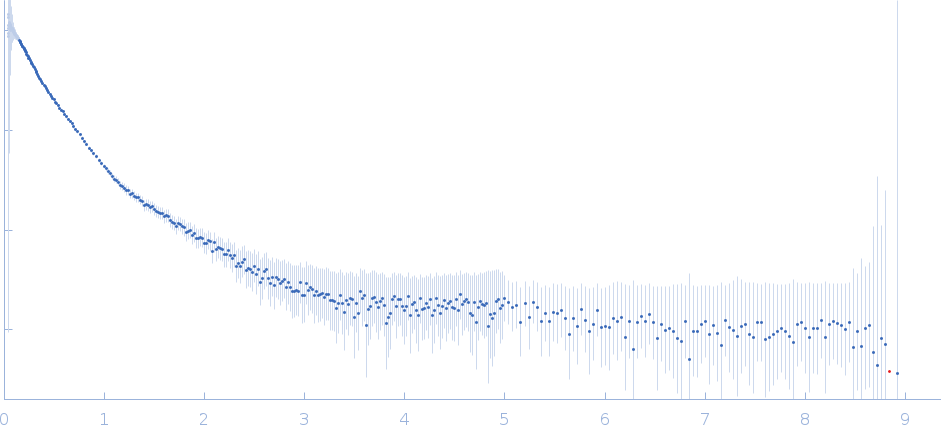
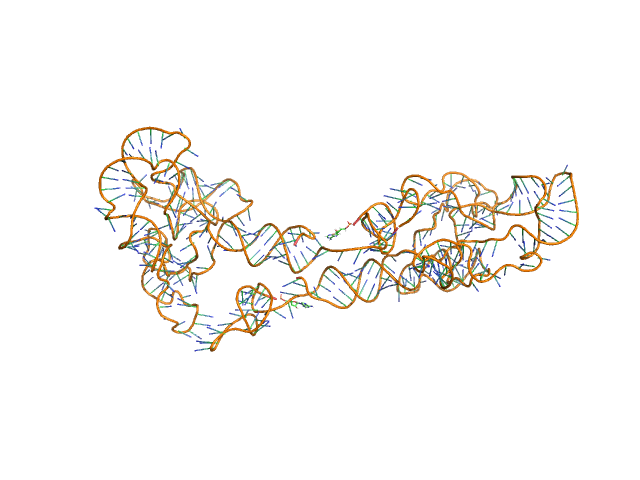
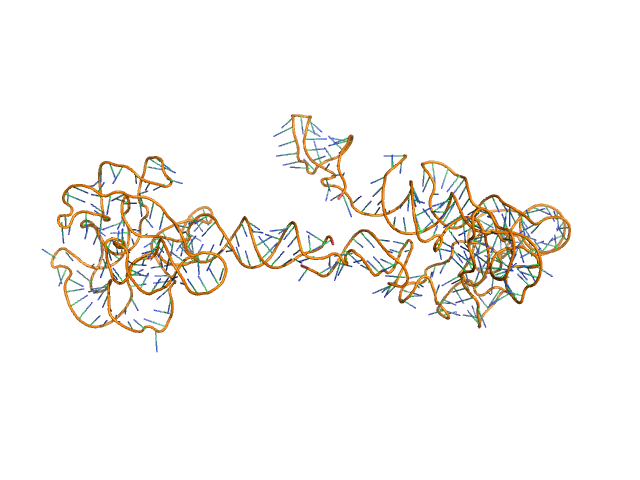
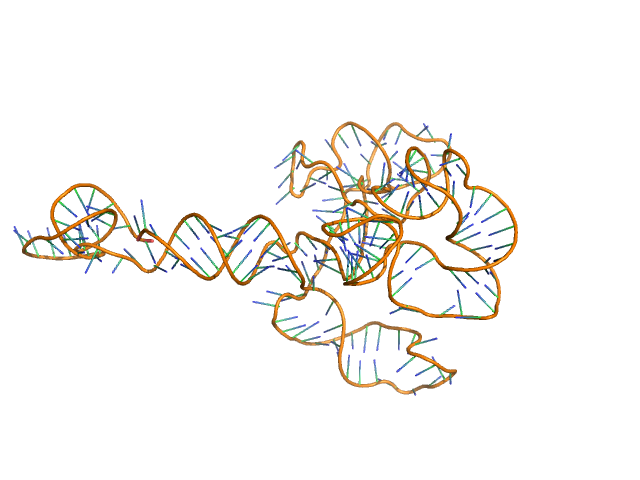
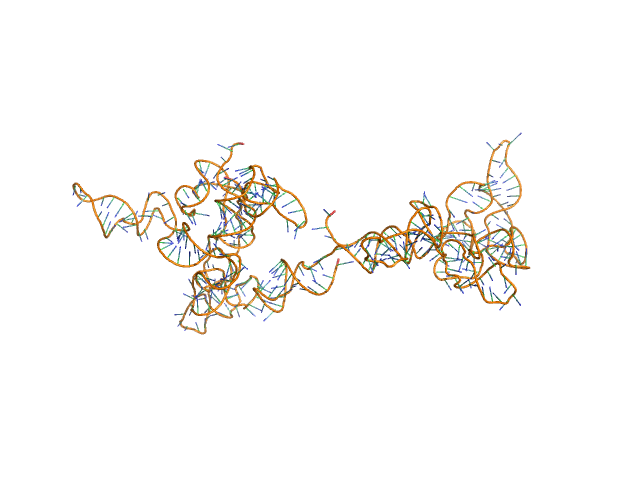
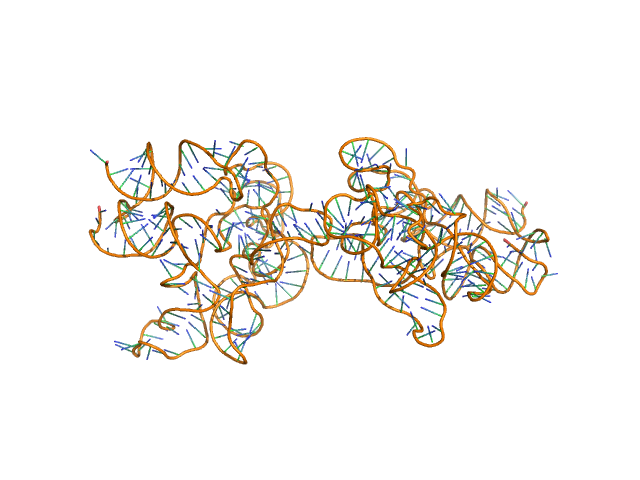
Synchrotron SAXS data from solutions of the fecB RNA riboswitch aptamer domain (rCbl) in 50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, 4-18 μM coenzyme B12 ligand, were collected on the 12ID-B SAXS/WAXS beam line at the Advanced Photon Source (APS; Argonne National Laboratory; Lemont, IL, USA) using a Pilatus 2M detector at a sample-detector distance of 1.9 m and at a wavelength of λ = 0.0932 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 0.07 mg/ml was measured at 10°C. 225 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
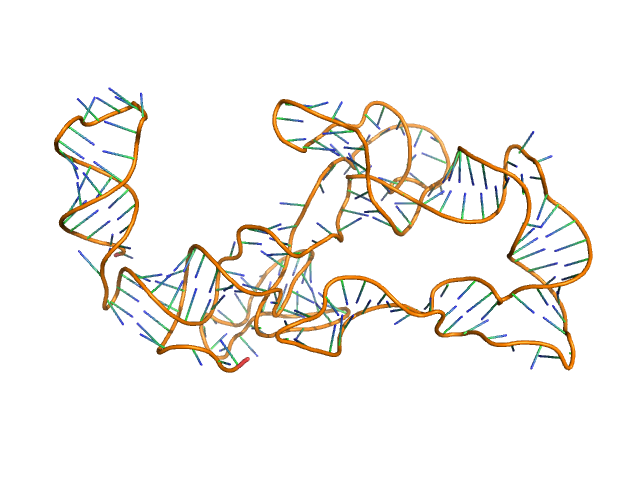
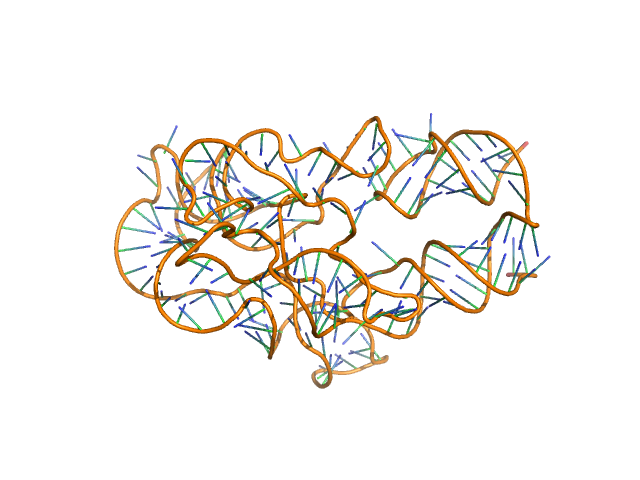
AFM single-particle visualization of rCbl in presence of ligands revealed a mixture of monomeric and dimeric species, each with various conformations. A few higher order oligomers were also observed. See the manuscript for detailed description of conforrmational and structural heterogeneity. The non- linearity of Guinier plot confirms this is not a monodisperse system. The Rg value may depend on the selection of the Guinier region. |
|
|||||||||||||||||||||