|
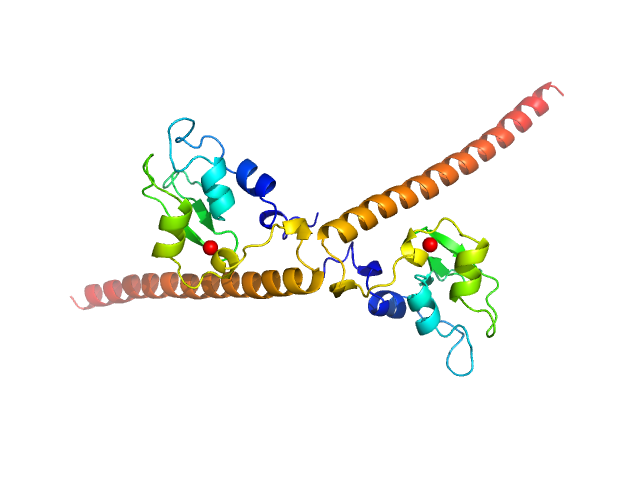
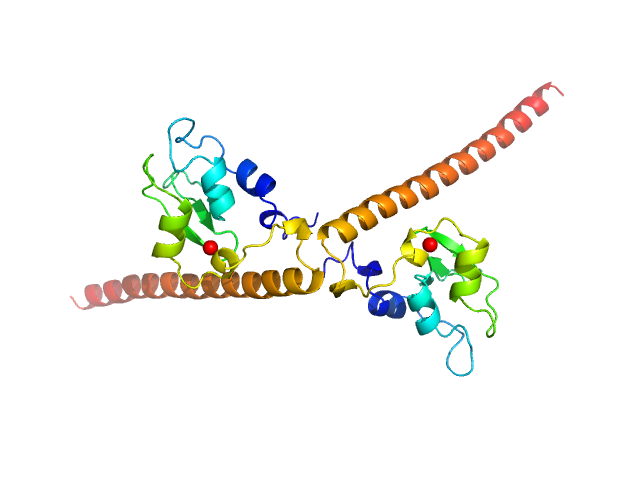
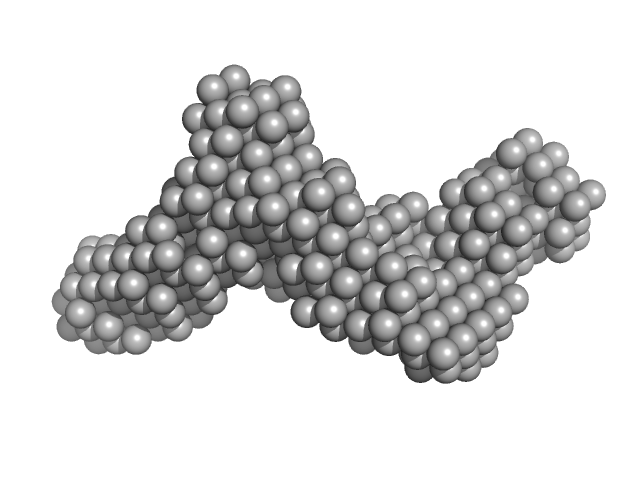
Synchrotron SAXS data from solutions of human survivin (BIRC5) in 50 mM Tris, 150 mM NaCl, 1 mM DTT, pH 8 were collected on the BM29 beam line at the ESRF (Grenoble, France) using a Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.099 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 500.00 μl sample at 3 mg/ml was injected at a 0.60 ml/min flow rate onto a GE Superdex 75 Increase 10/300 column at 10°C. 2500 successive 1 second frames were collected through the entire SEC column elution. Thirty-three frames from the sample were selected and processed (averaged and buffer subtracted) using CHROMIXS and primusqt from the ATSAS package version 3.0.0. The radius of gyration (Rg), forward scattering (I(0)), maximum particle dimension (Dmax) and the distance distribution function were determined using primusqt and GNOM. This molecule's calculated/expected molecular weight (MW) is around 16.77 kDa as a monomer and 33.54 kDa as a dimer. The MW estimation from the SAXS data (primusqt) from the different methods suggests a molecular weight of around 33 kDa, which fits well with the dimer. The models displayed in this entry included: Top/Middle: X-ray crystallography model (PDB ID: 6SHO) of the dimeric human Survivin compared with the experimental scattering curve using Crysol and FoXS server, respectively. Bottom: Single ab initio model obtained by dummy atom modelling using DAMMIF and refined with DAMMIN.
|
|
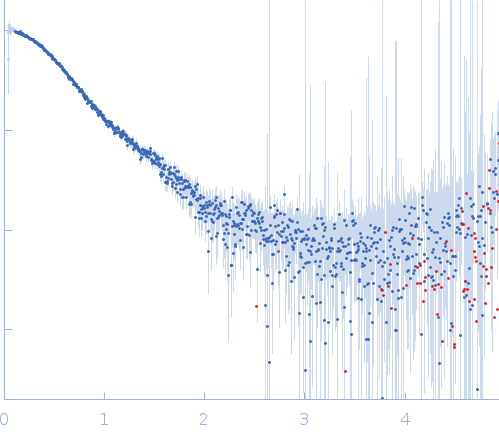 s, nm-1
s, nm-1