|
Synchrotron SAXS
data from solutions of
SH2-SH3-SH2 domains of Ras GTPase-activating protein 1 (p120RasGAP) bound to a doubly phosphorylated p190RhoGAP peptide
in
20 mM Tris pH 8, 350 mM NaCl, 1 mM DTT, pH 8
were collected
on the
BioCAT 18ID beam line
at the Advanced Photon Source (APS), Argonne National Laboratory storage ring
(Lemont, IL, USA)
using a Pilatus3 X 1M detector
at a sample-detector distance of 3.6 m and
at a wavelength of λ = 0.1003 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 320.00 μl sample
at 3.8 mg/ml was injected at a 0.50 ml/min flow rate
onto a GE Superdex 200 Increase 10/300 column
at 22°C.
1313 successive
0.500 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
On the day of data collection the samples were thawed and spun down. 1 mM DTT was added to the samples and buffer. The samples were injected onto a GE Superdex 200 Increase column at 22˚C in buffer containing 20 mM Tris pH 8, 350 mM NaCl, 1 mM DTT at a flow rate of 0.5 mL/min. Data were collected at APS’s BioCAT Beamline18ID using synchrotron radiation from Undulator A. As the sample came off of the column it was first detected by UV, followed by Wyatt DAWN HELEOS II MALS+DLS and Wyatt Optilab T-rEX dRI detectors for measuring molecular weight through MALS-DLS-RI analysis. Following MALS-DLS, the sample flowed to the SAXS sample chamber. The SAXS data were normalized using an active beamstop containing a silicon PIN diode.
|
|
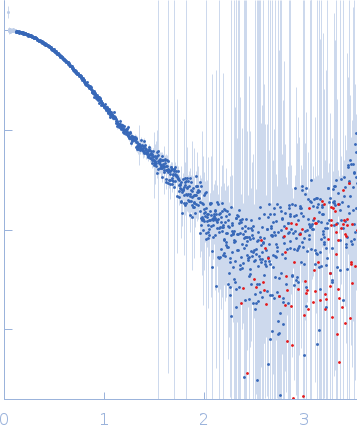 s, nm-1
s, nm-1