|
Synchrotron SAXS data from solutions of bovine serum albumin separated using asymmetric flow field flow fractionation (AF4) in 10 mM HEPES, 5 mM NaCl, 0.1 mM EDTA, pH 7.4 were collected on the EMBL P12 beam line at PETRA III (DESY, Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.155 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). The samples were fractionated by an asymmetrical flow field-flow fractionation (AF4) system (PN AF2000 MT). A semi-preparative frit-inlet AF4 (Fl-AF4) channel (shoulder width 50 mm, tip width 5 mm, tip-to-tip length 277 mm) was equipped with a polyethersulfone PES membrane with 10 kDa molecular weight cut-off and a Mylar spacer of 350 µm height. A solute concentration of 5 mg/ml was injected and separation was performed at at 20°C using a laminar flow. 2500 successive 1 second SAXS data frames were collected through the entire AF4 elution, where 23 sample data frames were selected through the monomer-separated elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
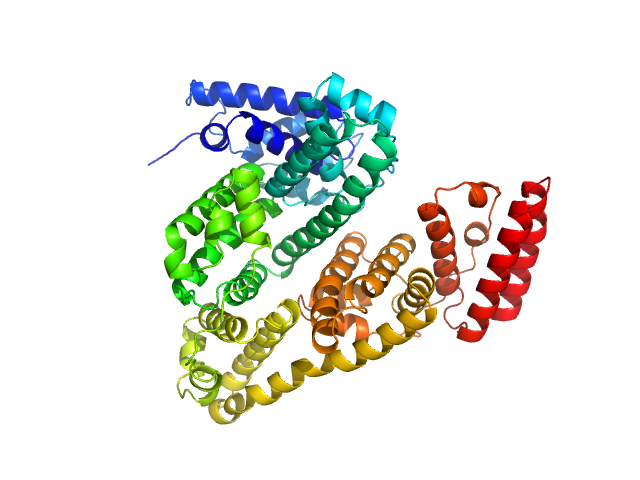
SAXS data frames corresponding to monomeric BSA were obtained with AF4-SAXS and displayed a good fit to the calculated scattering curve derived from the atomistic X-ray crystal structure. The comparison to the data collected in static mode clearly shows that larger species were removed from the sample through the separation process. The decrease in scattering at low-s along with the decrease in radius of gyration, Rg (from 3.4 ± 0.1 nm to 2.9 ± 0.1 nm ), maximum distance, Dmax (from 13 ± 0.4 nm to 9.1 ± 0.2 nm ), and the molecular weight estimate (based on the volume of correlation from 82 ± 8 kDa to 69 ± 7 kDa) are all in line with the scattering of monomeric BSA as opposed to the mixture containing roughly 10-20% dimeric and larger oligomeric fractions.
|
|
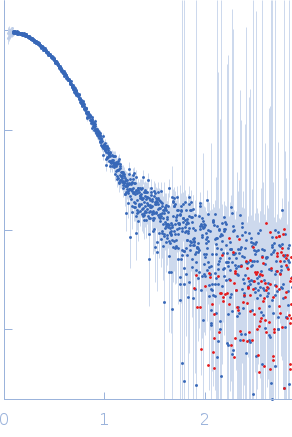 s, nm-1
s, nm-1