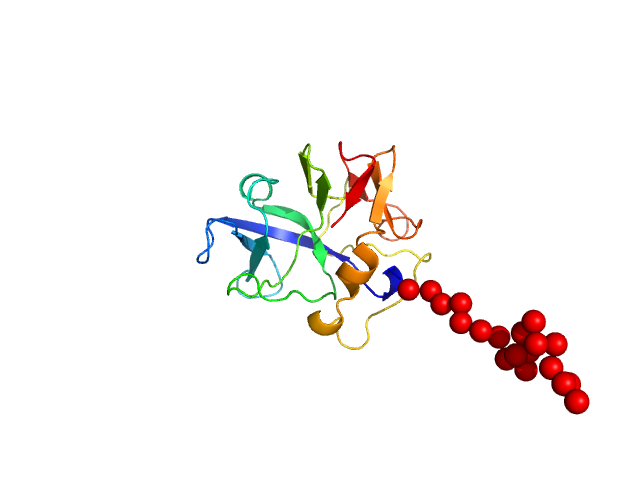
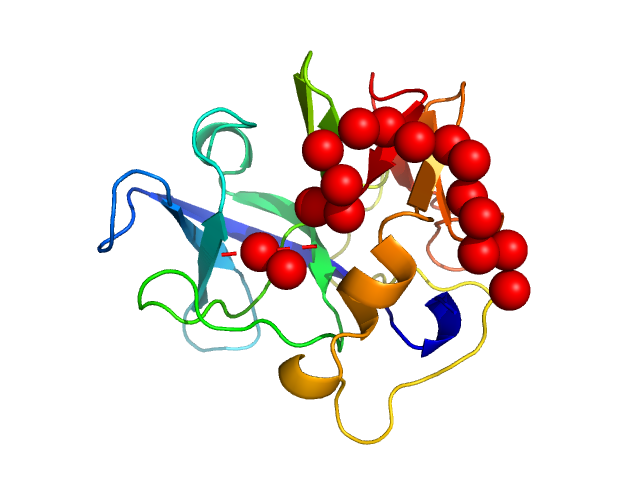
|
SAXS data from the solutions of EMAPII at 4 mg/ml concentratio in 50 mM Tris pH8, 150 mM NaCl, 1 mM DTT were collected using Xenocs Xeuss 2.0 System (Department of Biomedical Physics, Adam Mickiewicz University, Poznań, Poland) equipped with a MetalJet D2 microfocus X-ray generator and a Pilatus 3R 1M detector at a wavelength of λ = 0.134 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Before SAXS experiments protein sample was purified using SEC chromatography and concentrated to 4 mg/ml using Amicon Ultra 4, 3k centrifugal filter. Protein sample at 4.00 mg/ml concentration was measured at 20°C. 6x600 second frames were collected. The data were radially averaged and the scattering of the solvent was subtracted. Modeling of the EMAPII protein was performed using Ensemble Optimization Method (EOM) using NMR structure of the EMAPII protein (PDB ID: 8ONG, Entry Title: Structure of the endothelial monocyte activating polypeptide II (EMAP II) in solution) where first 19 amino acids where represented as dummy residues.
Cell temperature = UNKNOWN
|
|
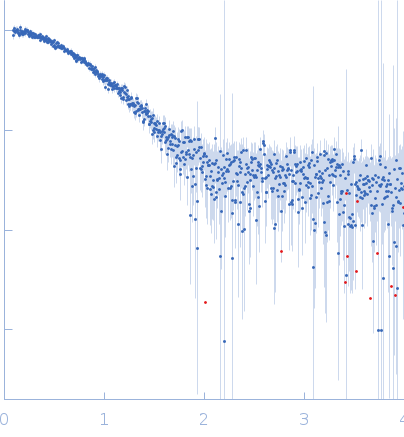 s, nm-1
s, nm-1
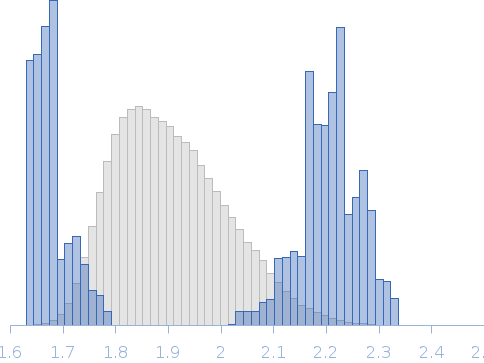 Rg, nm
Rg, nm