|
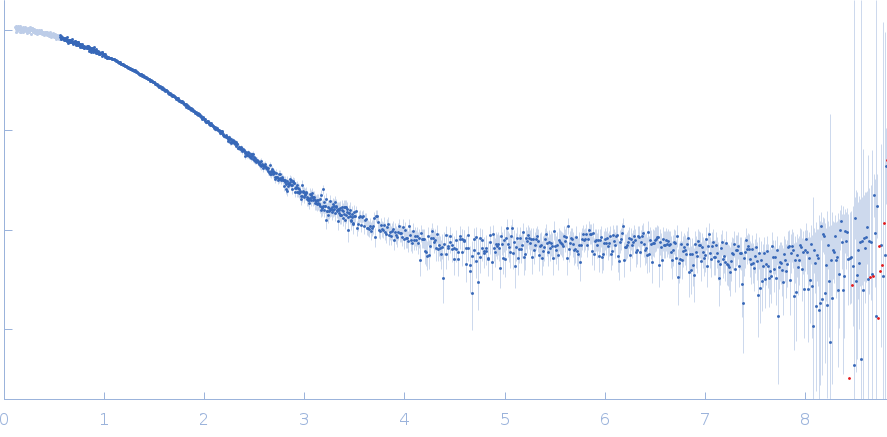
Synchrotron SAXS data from solutions of poly(GU) RNA with 12 GU repeats in 20 mM HEPES, 150 mM KCl, pH 7 were collected on the 12-ID-B beam line at the Advanced Photon Source (APS), Argonne National Laboratory (Lemont, IL, USA) using a Eiger 2S detector at a sample-detector distance of 1.9 m and at a wavelength of λ = 0.0932 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 0.3 and 1.6 mg/ml were measured at 22°C. 60 successive 0.500 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. The scattering curves through the concentration series, 0.26, 1, 1.4 and 1.6 mg/ml, were extrapolated to infinite dilution to obtain the final curve displayed in this entry.
|
|
 s, nm-1
s, nm-1
