|
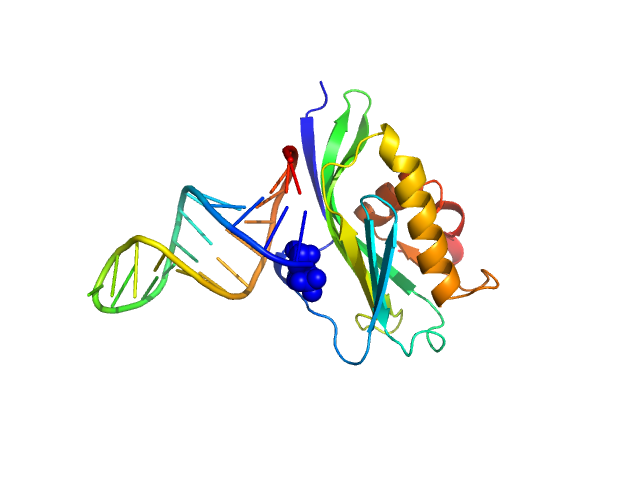
Samples of NS1B-CTD* were prepared at a concentration of 250 μΜ (4 mg/mL) and 1:1 ratio of NS1B-CTD* : 5’ppp ds10 HP RNA in 1K buffer (40 mM ammonium acetate, pH 5.5, 225 mM NaCl, 5 mM CaCl2, 0.02% NaN3, 50 mM arginine) as this lower salt buffer provides a tighter complex. Immediately before mixing the protein with RNA to make the complex, NS1B-CTD* was buffer exchanged into 1K buffer, and concentrated to provide a 250 μΜ complex concentration after adding RNA. RNA was then added to provide a 1:1 NS1B-CTD : 5’ppp ds10 HP RNA ratio. Samples were centrifuged at 8,500 x g for a minimum of 10 mins at the beamline immediately before data collection on the supernatant. SAXS was performed at BioCAT beamline 18ID (Advanced Photon Source, Chicago, IL). Data were collected in a SAXS flow cell which consists of a 1.5 mm ID quartz capillary with 10 µm walls held at 20 oC using 12 keV incident X-rays. Scattering intensity was recorded using a Pilatus3 X 1M (Dectris) detector which was placed 3.44 m from the sample, providing access to a s-range of 0.12 nm-1 to 3 nm-1. During exposure, the sample was flowed through the beam in a single direction, and 0.5 s exposures were acquired every 2 seconds during flow. Data was reduced using BioXTAS RAW 1.4.1. Buffer blanks were created by recording SAXS data from a matched buffer and subtracted from exposures from the sample to create the I(s) vs s curves used for subsequent analyses. All sets of nominally identical measured profiles were automatically compared using CorMap to test for radiation damage or other changes. SAXS data quality assessed by Guinier plot analysis. These data indicated little or no aggregation of the complex under these conditions, consistency between expected and observed molecular weight, low sample radiation damage, and appropriate P(r) calculations.
|
|
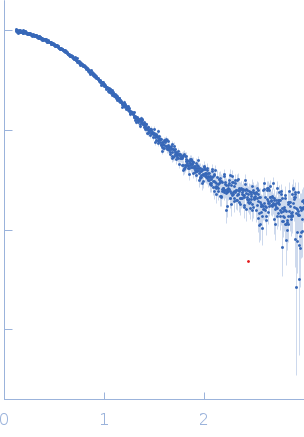 s, nm-1
s, nm-1