|
Data was collected at the SAXS/WAXS beam line at the Australian Synchrotron (Melbourne, Australia) using a Pilatus3 S 2M detector at a sample-detector distance of 2.68 m and at a wavelength of λ = 0.107812 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 50 μl sample at 2.587 mg/ml was injected onto a GE Superdex 200 5/150 column at 25°C using 150 mM KCl, 20 mM HEPES (pH 7.4), 5 mM MgCl2, 1 mM DTT, 5% glycerol buffer. 400 successive 1 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
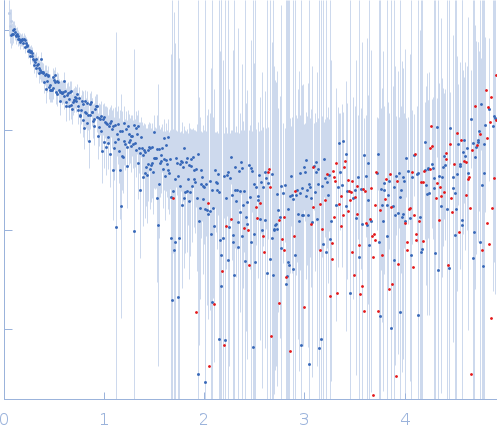
This is the 60 bp GAGE6 oligo from Lee et al (2015); Wang et al (2022) which reportedly binds SFPQ. SAXS analysis indicates some degree of curvature which is likely due to the presence of multiple AT repeat regions in the sequence.
|
|
 s, nm-1
s, nm-1
