|
Synchrotron SAXS data from solutions of Surface glycan binding protein (Cf-SGBP) from Christiangramia forsetii in 10 mM MOPS, 100 mM NaCl, pH 7.8 were collected on the SWING beam line at the SOLEIL storage ring (Saint-Aubin, France) using a AVIEX PCCD170170 detector at a sample-detector distance of 2.4 m and at a wavelength of λ = 1.033 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle) resulting in a scattering vector q-range of 0.012 – 0.504 Å-1.
In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 70.00 μl sample at 8.2 mg/ml was injected at a 0.20 ml/min flow rate onto a Agilent Bio SEC-5, 500Å column at 17°C. 250 successive 1.500 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
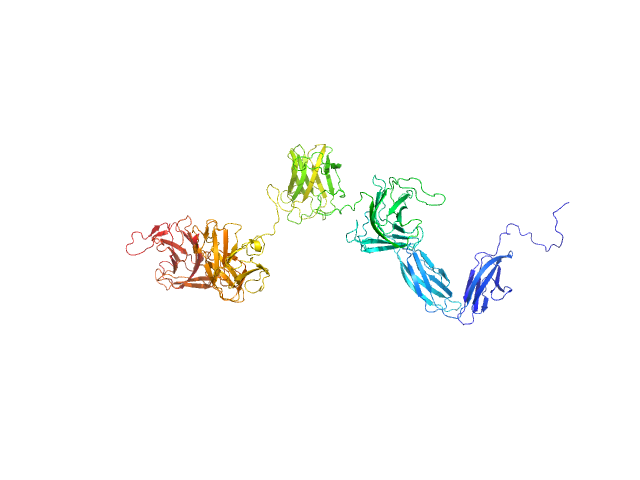
The protein construct used for SAXS contains a N-terminal polyhistidine tag MGSSHHHHHHGS.
Protein samples were centrifuged prior to analyses to remove putative aggregates. SAXS measurements were coupled with prefixed SEC. The SEC column was equilibrated in buffer C (100 mM NaCl, 10 mM MOPS pH 7.8), which was used for protein purification.
The Guinier equation was used to calculate the forward scattering I(0) and the radius of gyration Rg. The distance distribution function P(r) and the maximum particle dimension Dmax were calculated by Fourier inversion of the scattering intensity I(q) using GNOM integrated in the PRIMUS software (ATSAS 3.1.3).
|
|
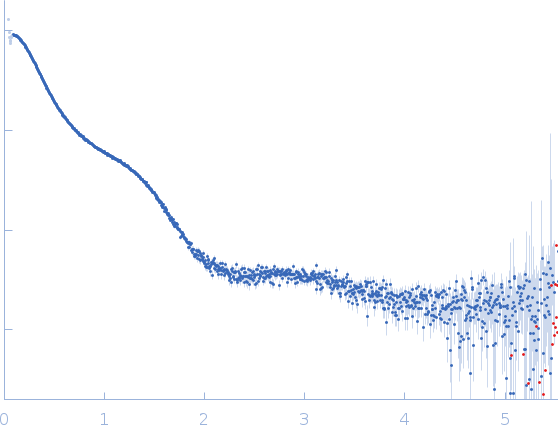 s, nm-1
s, nm-1