|
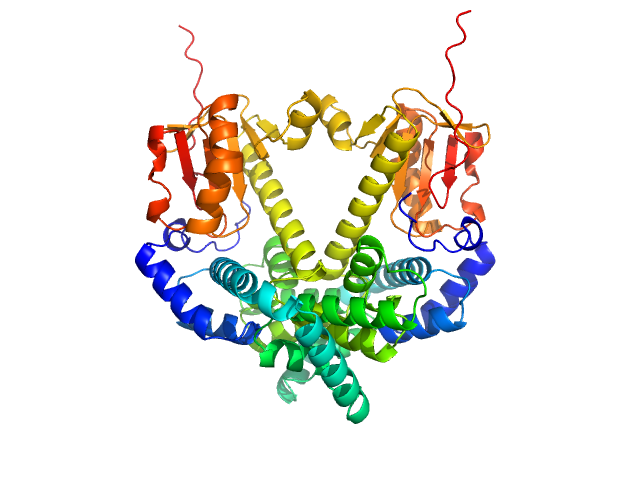
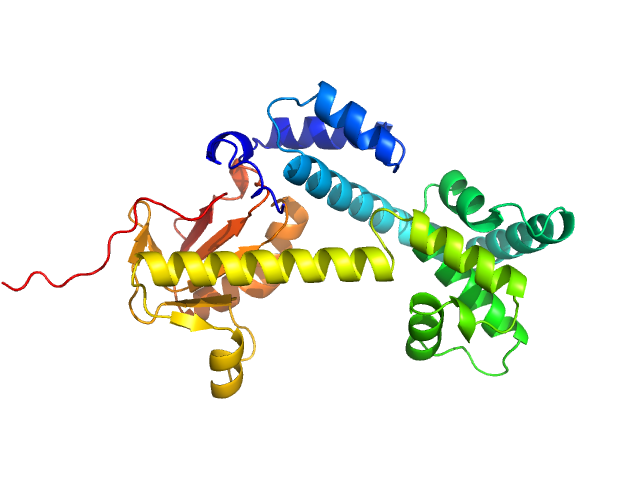
The SEC-SAXS data from the solution of CDIN1 diluted in 20 mM Tris HCl, 150 mM NaCl, 5% glycerol, pH 8.0 buffer were collected on the EMBL P12 beamline at the PETRA III storage ring (DESY; Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.123982 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAXS was employed. The sample (45 μl at 10 mg/ml) was injected and ran into a GE Superdex 200 Increase 5/150 column at a flow rate of 0.3 ml/min at 20°C. Scattering data were collected throughout the entire SEC column elution with an exposure time of 0.495 sec/frame (for 1200 frames total). The data were normalized to the intensity of the transmitted beam. The selected area of sample elution peak was selected and processed (averaged and buffer subtracted) from 10 data frames using CHROMIXS and primusqt from the ATSAS 3.2.1 package. The radius of gyration (Rg), forward scattering (I(0)), maximum particle dimension (Dmax), and the distance distribution function were determined using primusqt and GNOM. The calculated molecular weight (MW) of CDIN1 is around 67 kDa. MALDI-TOF mass spectrometry suggests a MW 33 369 Da for the CDIN1 monomer. The MW estimation from the SAXS data (primusqt) varied depending on the method used: Qp = 66.3 kDa, MoW = 62.3 kDa, Vc = 66.6 kDa, Size & Shape = 76.6 kDa (Bayesian Inference MW, 62.3 kDa in the 92% credibility interval of 58.8-67.9 kDa). The models displayed in this entry included: Top: The Alphafold2 predicted model of CDIN1 dimer compared with the SAXS data using CRYSOL. Bottom: The Alphafold2 predicted model of CDIN1 monomer compared with the SAXS data using CRYSOL.
|
|
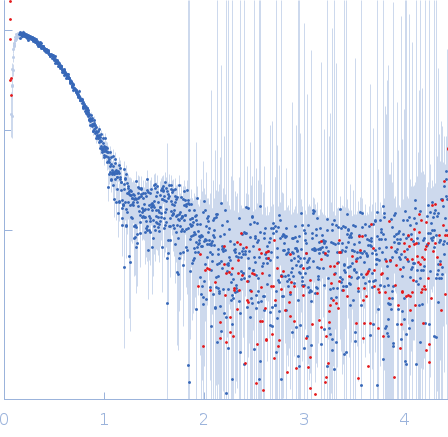 s, nm-1
s, nm-1