|
Synchrotron SAXS data from solutions of Apo FphH in 100 mM NaCl, 10 mM HEPES, pH 7.6 were collected on the BioSAXS beam line at the Australian Synchrotron (Melbourne, Australia) using a Pilatus3 S 2M detector at a sample-detector distance of 2.5 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 40.00 μl sample at 5.4 mg/ml was injected at a 0.40 ml/min flow rate onto a GE Superdex 75 Increase 5/150 column at 21°C. 16 successive 0.500 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
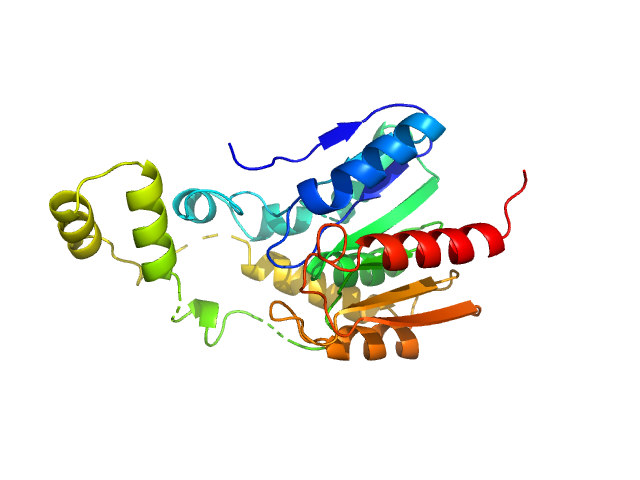
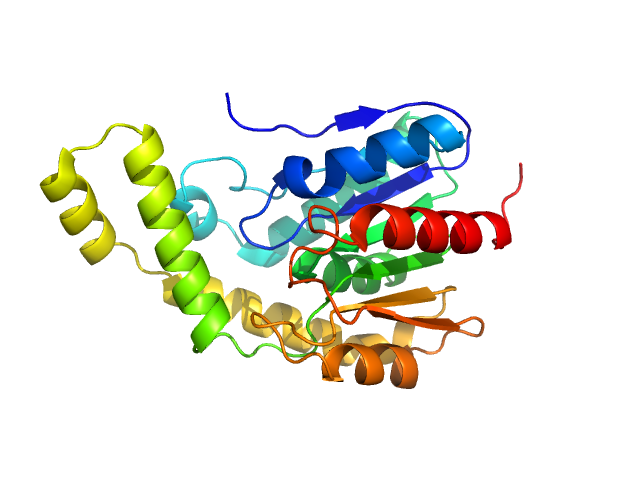
A serine hydrolase with short chain esterase activity, active in biofilm life cycle of S. aureus. Two shape reconstructions are displayed: 1) DAMFILT (top) derived from the volume and bead occupancy corrected spatial alignment of several individual DAMMIF models and; 2) An individual DAMMIF model example (bottom) with the corresponding fit to the data shown to the left.
|
|
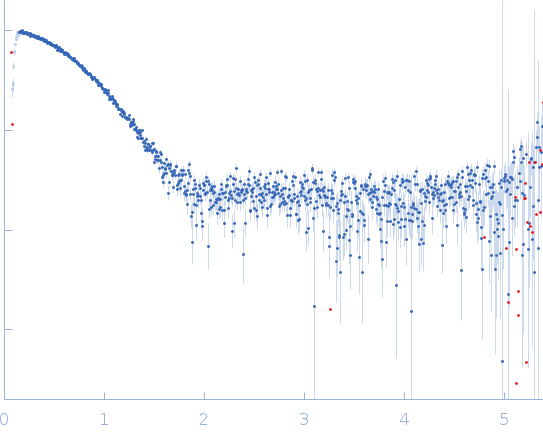 s, nm-1
s, nm-1