|
Synchrotron SAXS
data from solutions of
N-acetylmuramoyl-L-alanine amidase (with His-Tag)
in
20 mM Tris, 150 mM NaCl and 1 mM DTT, pH 8
were collected
on the
B21 beam line
at the Diamond Light Source storage ring
(Didcot, UK)
using a Eiger 4M detector
at a sample-detector distance of 3.7 m and
at a wavelength of λ = 0.1 nm
(I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle).
One solute concentration of 5.60 mg/ml was measured
at 18°C.
600 successive
3 second frames were collected.
The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
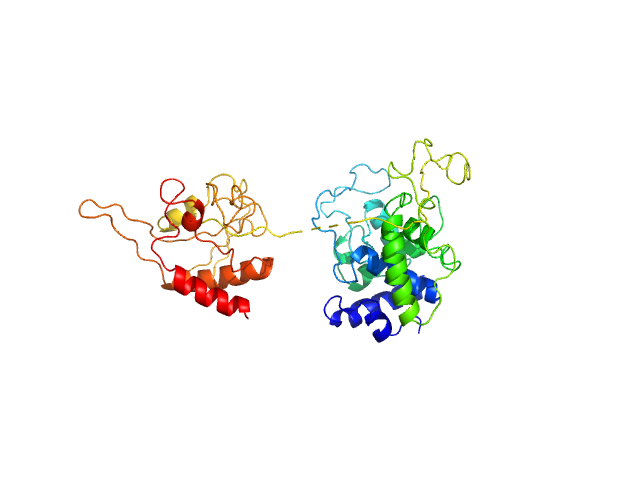
SAXS experiments were performed at the beamline B21 of the Diamond Light Source (Harwell Campus, UK) (Cowieson et al, 2020). Samples of 45 µl of 6His-tagged and untagged DS6AGH19-Ami2B protein at 5.6 and 5.9 mg/ml were loaded into a Sodex KW-403 column and a SRT-C SEC-300 (Sepax) column, respectively, equilibrated in buffer (20 mM Tris pH 8.0, 150 mM NaCl and 1 mM DTT) and connected to an Agilent 1200 HPLC system at 18 °C. Continuously eluting samples were exposed for 3 s in 10 s acquisition blocks using an X-ray wavelength of 1 Å, and a sample to detector (Eiger 4M) distance of 3.7 m. The data covered a momentum transfer range of 0.0032 < q < 0.34 Å−1. The frames recorded immediately before elution of the sample were subtracted from the protein scattering profiles. The Scåtter software package (www.bioisis.net) was used to analyse data, buffer-subtraction, scaling, merging and checking possible radiation damage of the samples. The Rg value was calculated with the Guinier approximation assuming that at very small angles q < 1.3/Rg. The particle distance distribution, Dmax, was calculated from the scattering pattern with GNOM, and shape estimation was carried out with DAMMIF/DAMMIN, all these programs included in the ATSAS package. The proteins molecular mass was estimated with GNOM. Interactively generated PDB-based homology models were made using the program COOT by manually adjusting the X-ray structures obtained in this work, into the envelope given by SAXS until a good correlation between the real-space scattering profile calculated for the homology model matched the experimental scattering data. This was computed with the program FoXS.
|
|
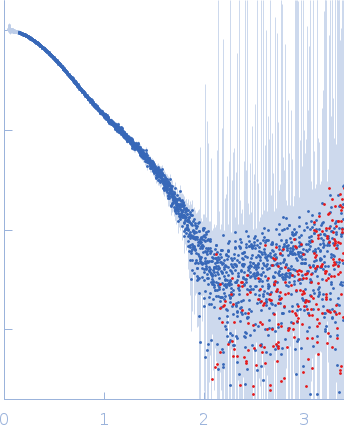 s, nm-1
s, nm-1