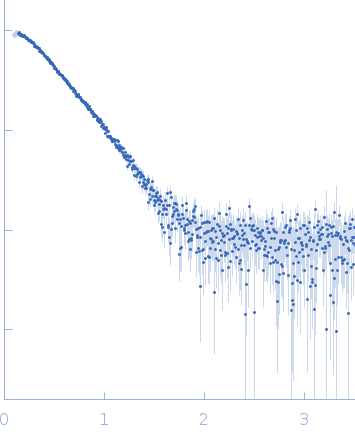
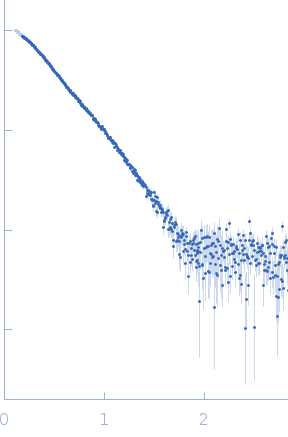
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Semrau MS,
Giachin G,
Covaceuszach S,
Cassetta A,
Demitri N,
Storici P,
Lolli G
Nat Commun
13(1):6199
(2022 Oct 19)
|
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|