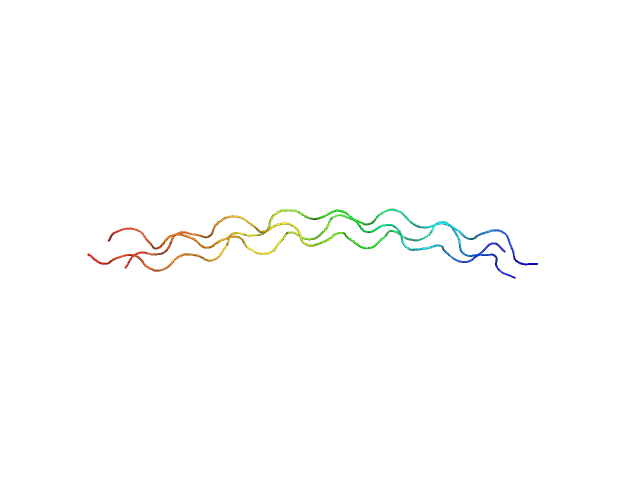A solution structure analysis reveals a bent collagen triple helix in the complement activation recognition molecule mannan-binding lectin.
Iqbal H,
Fung KW,
Gor J,
Bishop AC,
Makhatadze GI,
Brodsky B,
Perkins SJ
J Biol Chem
299(2):102799
(2023 Feb)
|
|
|
|

|
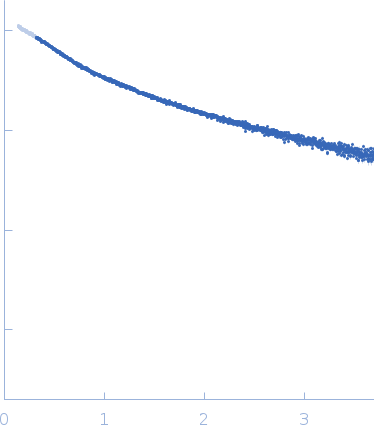
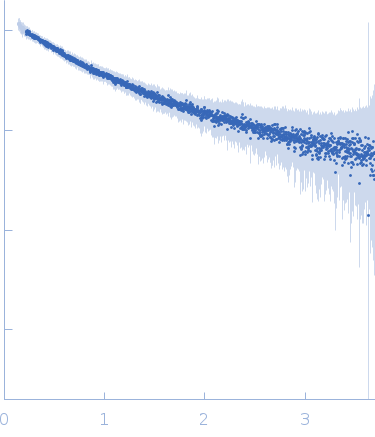
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
|
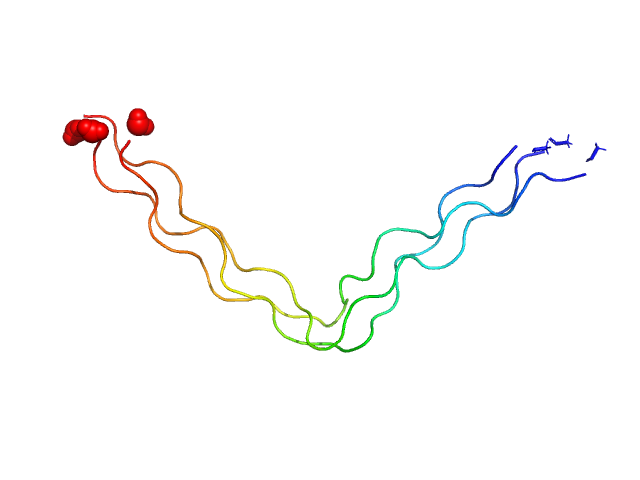
| Sample: |
Ac-(POG)4-QG-(POG)5-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)5-EOGQGLRG-(POG)3-NH2 trimer, 12 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
6 |
nm3 |
|
|
|
|
|
|
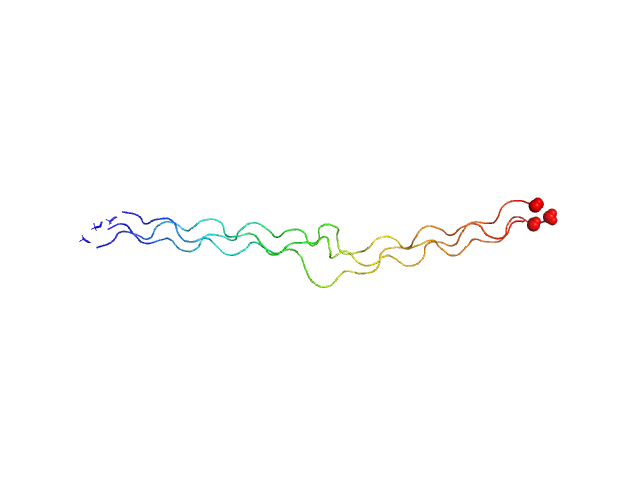
|
| Sample: |
Ac-(POG)5-EPGQGLRG-(POG)5-NH2 trimer, 14 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ac-(POG)10-NH2 trimer, 12 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
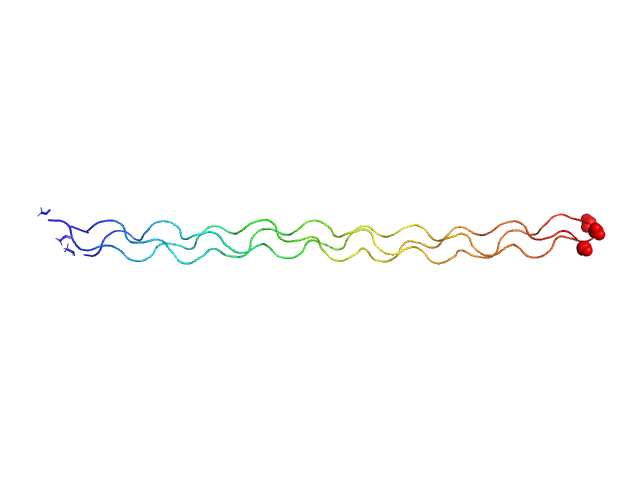
|
| Sample: |
Ac-(POG)13-NH2 trimer, 15 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
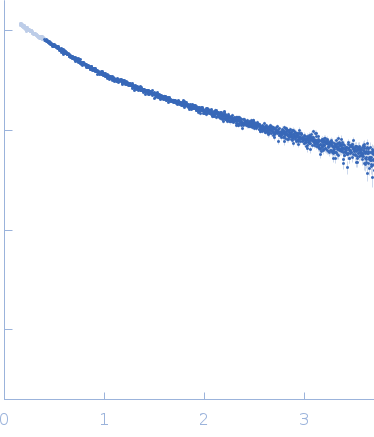
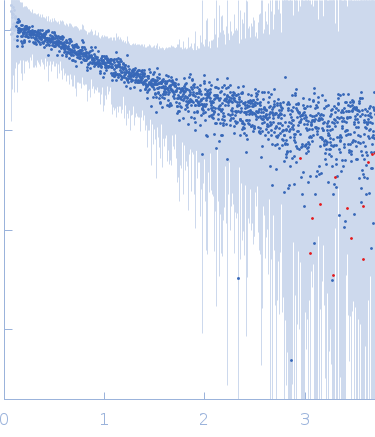
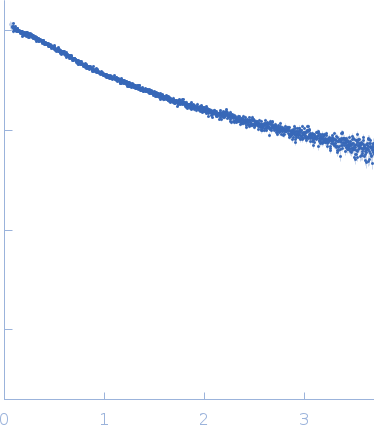
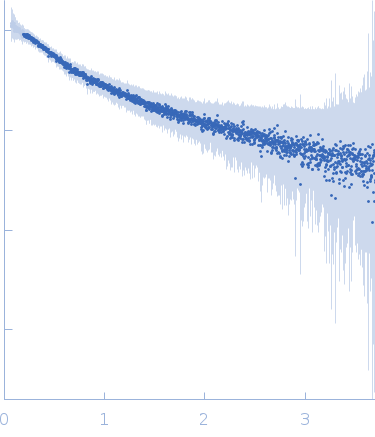
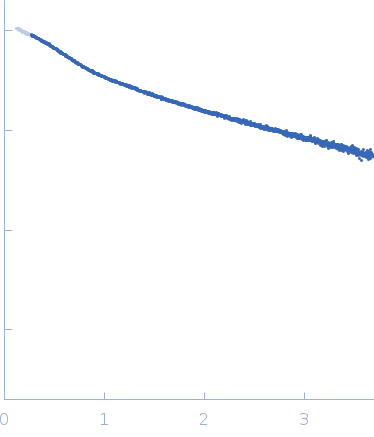
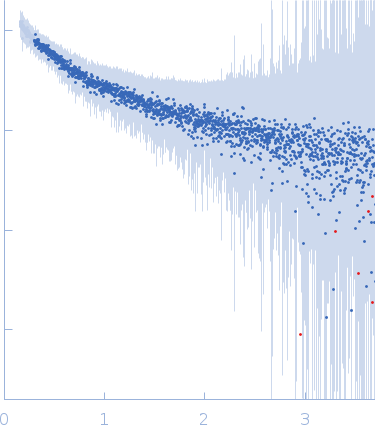
| Sample: |
[(Pro-Pro-Gly)10]3 trimer, 8 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)4-POA-(POG)5-NH2 trimer, 12 kDa protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ac-(POG)3-ITGARGLAG-(POG)4-NH2 trimer, 11 kDa synthetic construct protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.7 mM KCL,, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 1
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
9 |
nm3 |
|
|




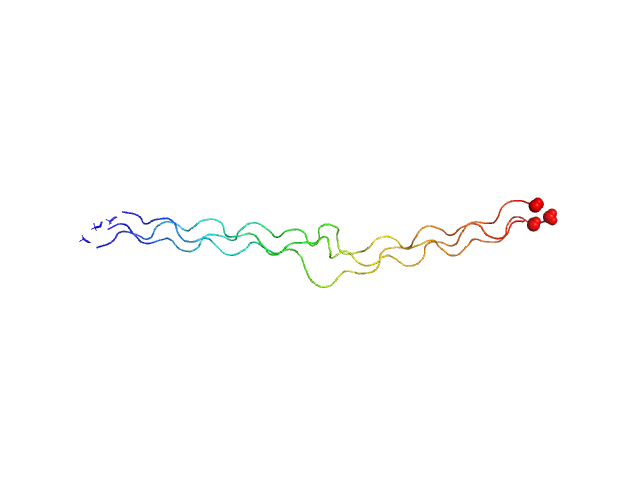
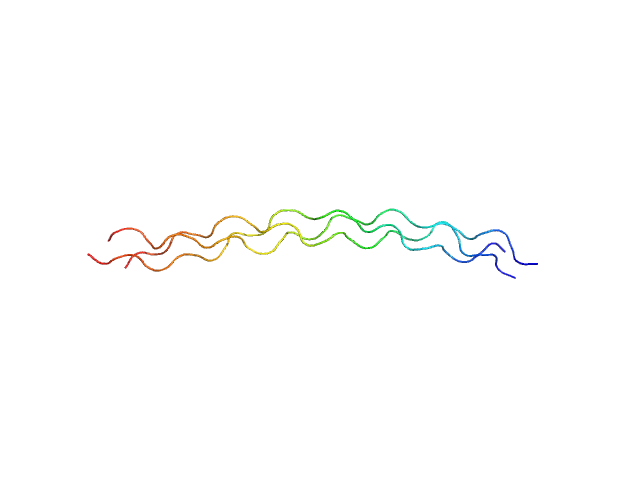
![[(Pro-Pro-Gly)10]3 experimental SAS data [(Pro-Pro-Gly)10]3 experimental SAS data](/media/intensities_files/scattering_plots/SASDRY2_dat_img.png)