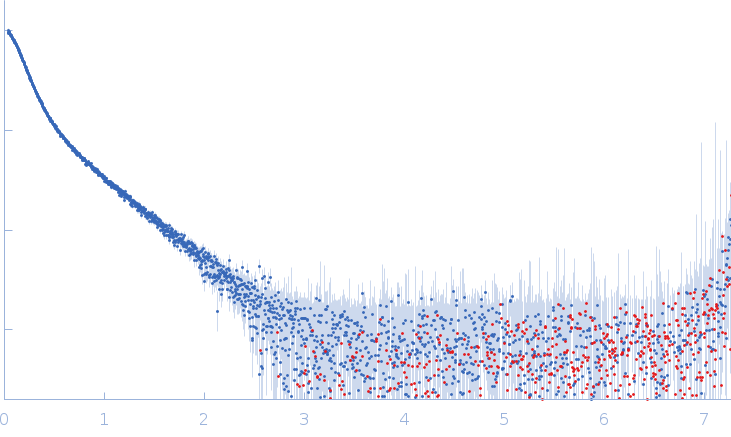
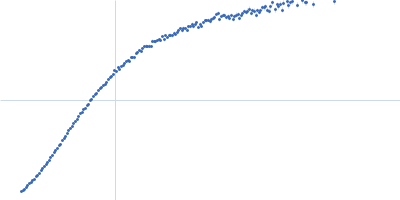
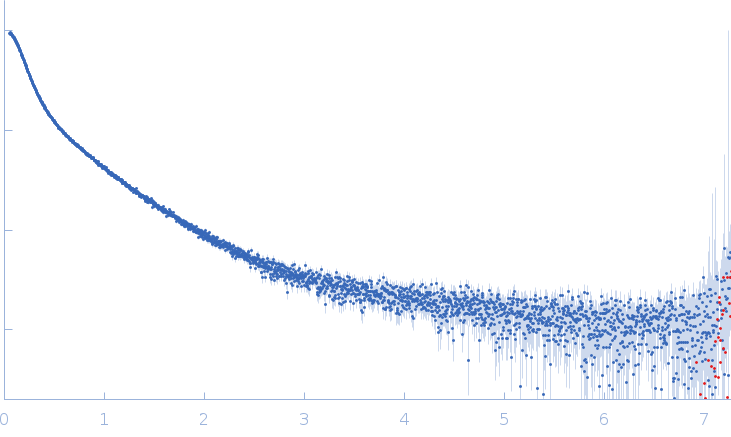
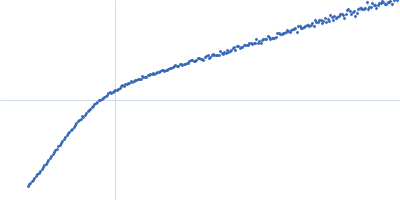
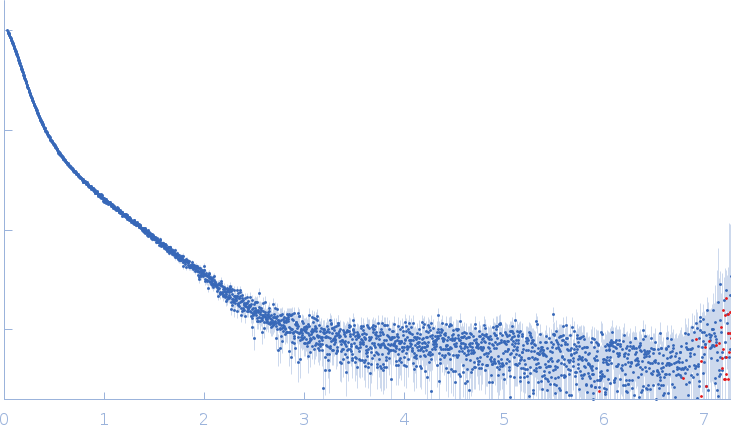
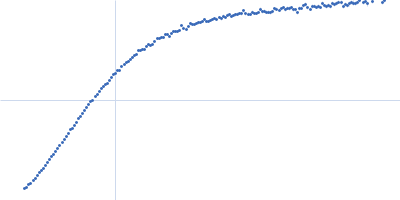
Conformation and structural dynamics of the Xist lncRNA A-repeats
Jones A,
Gabel F,
Bohn S,
Wolfe G,
Sattler M
(2023 Jul 31)
|
|
|
|
|
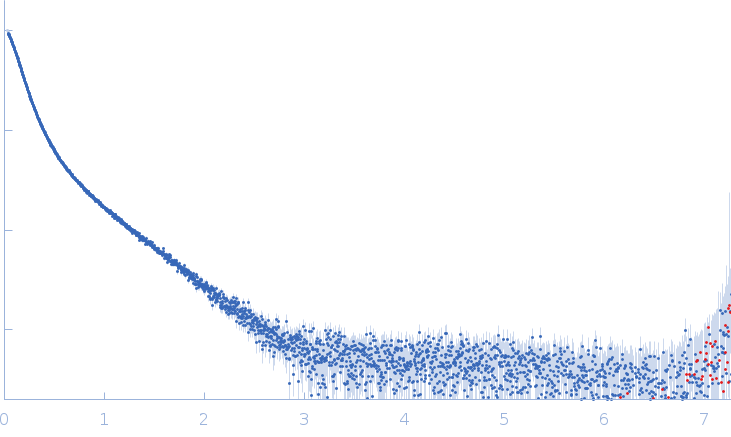
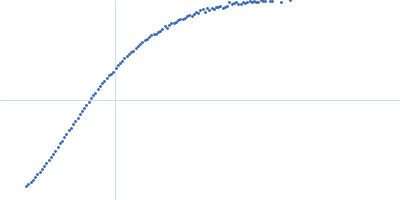
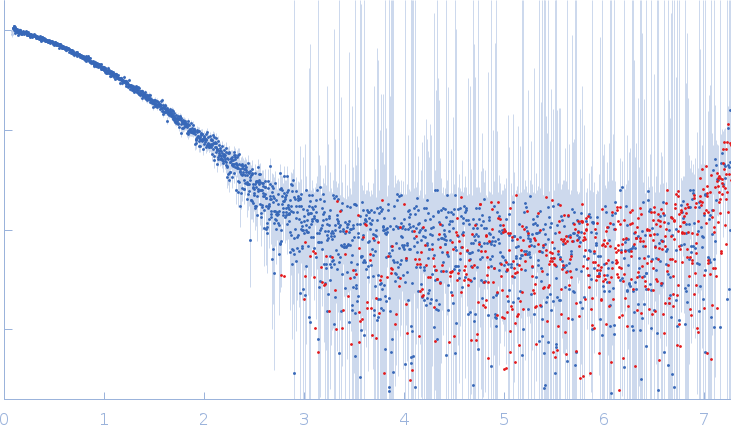
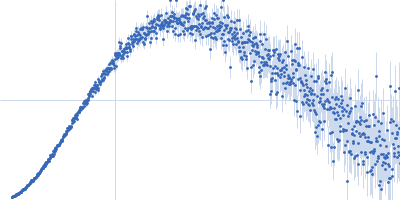
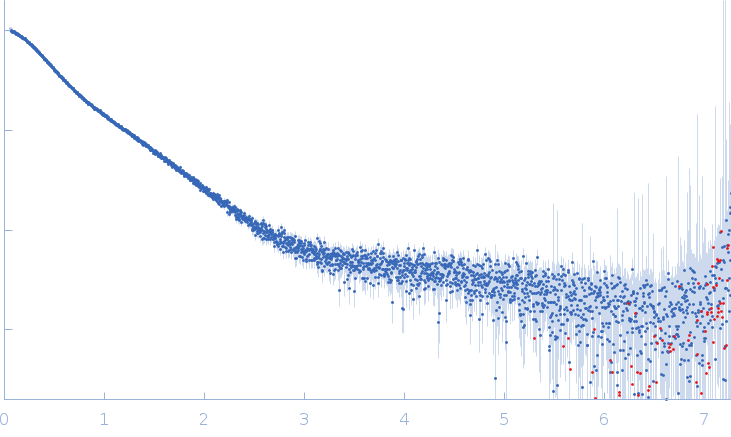
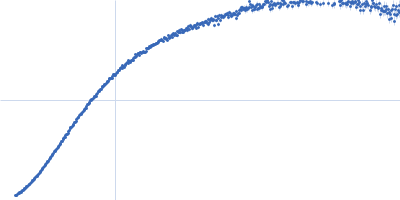
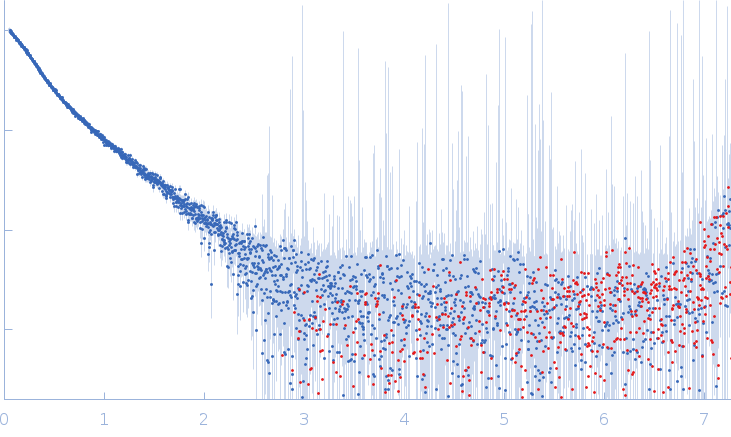
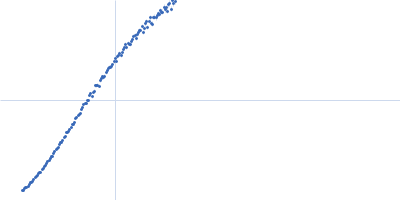
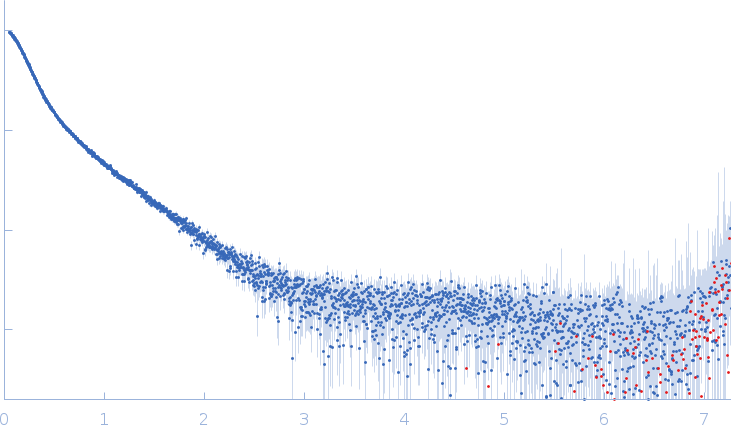
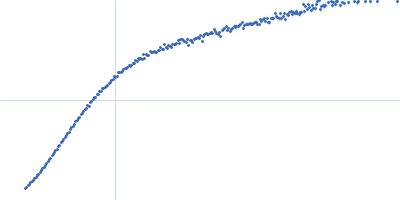
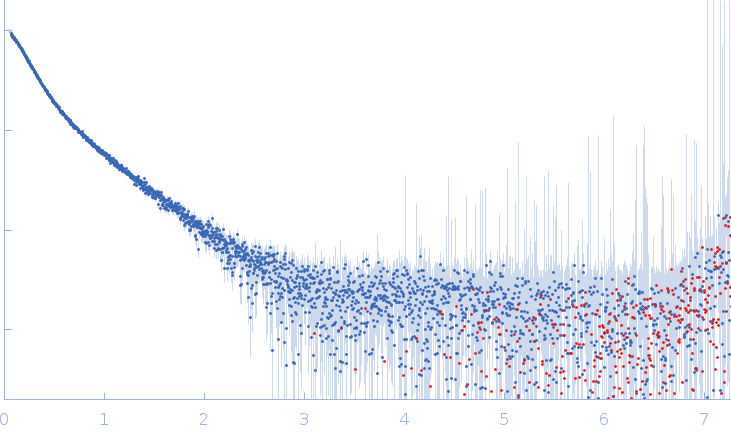
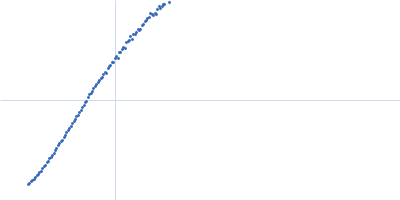
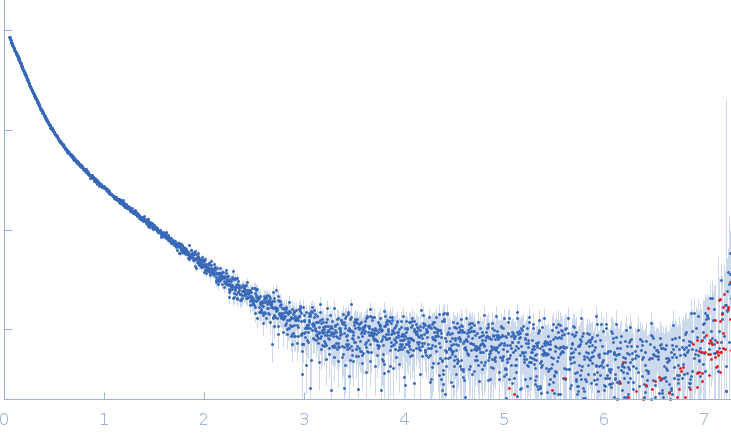
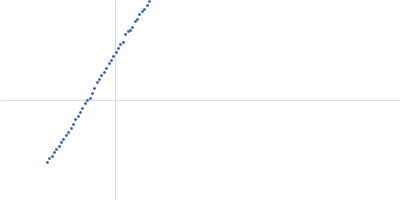
| Sample: |
Xist A-repeat lncRNA monomer, 148 kDa Homo sapiens RNA
|
| Buffer: |
25 mM K-HEPES, 0.1 mM Na-EDTA, 150 mM KCl, 15 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
12.1 |
nm |
| Dmax |
45.0 |
nm |
|
|
|
|
|
|
|
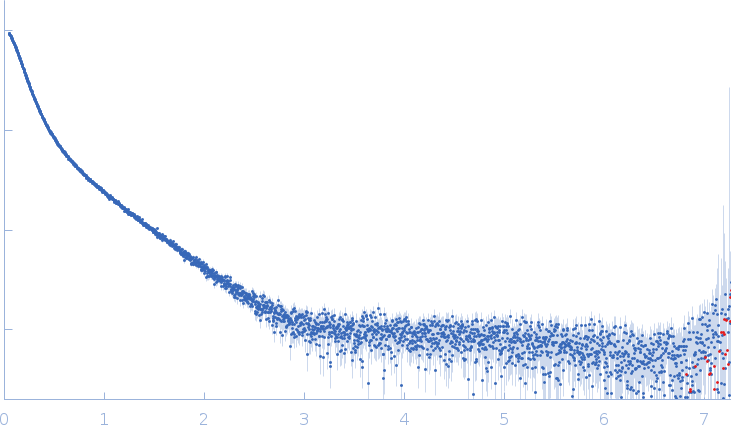
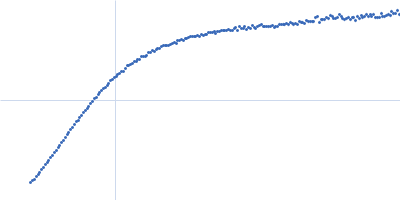
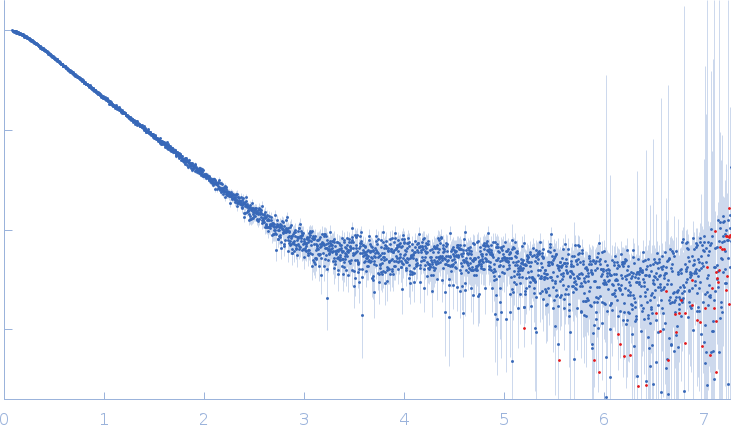
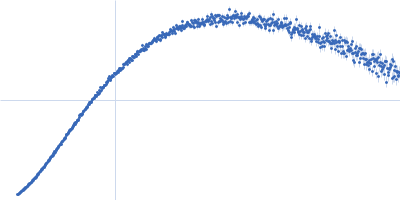
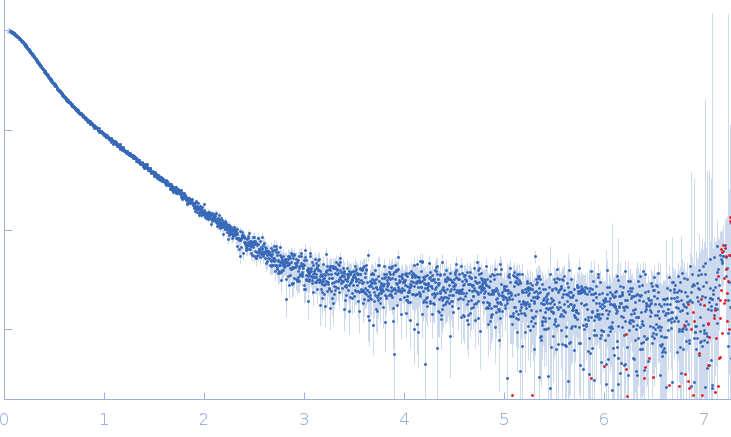
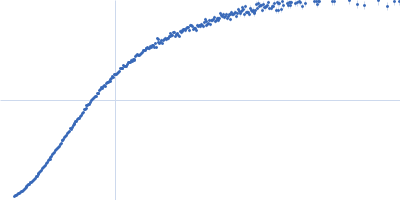
| Sample: |
Xist A-repeat lncRNA monomer, 148 kDa Homo sapiens RNA
|
| Buffer: |
100 mM HEPES, 100 mM NaCl, 10 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
9.8 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
657 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA monomer, 148 kDa Homo sapiens RNA
|
| Buffer: |
20 mM HEPES-KOH, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 20% glycerol, 3.25 mM MgCl2, pH: 7.9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
8.9 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
414 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA monomer, 148 kDa Homo sapiens RNA
|
| Buffer: |
10 mM NaH2PO4/Na2HPO4, 100 mM NaCl, 0.02 mM EDTA, 0.02% azide, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
7.6 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 14 mer monomer, 5 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 24 mer dimer, 16 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 1 repeat dimer, 41 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 2 repeats monomer, 34 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
5.0 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 3 repeats monomer, 48 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
6.3 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 4 repeats monomer, 62 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
7.2 |
nm |
| Dmax |
34.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 5 repeats monomer, 81 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 6 repeats monomer, 96 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
12.9 |
nm |
| Dmax |
38.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA monomer, 148 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
|
| RgGuinier |
11.2 |
nm |
| Dmax |
50.0 |
nm |
|
|