The structure of pathogenic huntingtin exon 1 defines the bases of its aggregation propensity.
Elena-Real CA,
Sagar A,
Urbanek A,
Popovic M,
Morató A,
Estaña A,
Fournet A,
Doucet C,
Lund XL,
Shi ZD,
Costa L,
Thureau A,
Allemand F,
Swenson RE,
Milhiet PE,
Crehuet R,
Barducci A,
Cortés J,
Sinnaeve D,
Sibille N,
Bernadó P
Nat Struct Mol Biol
(2023 Mar 2)
|
|
|
|
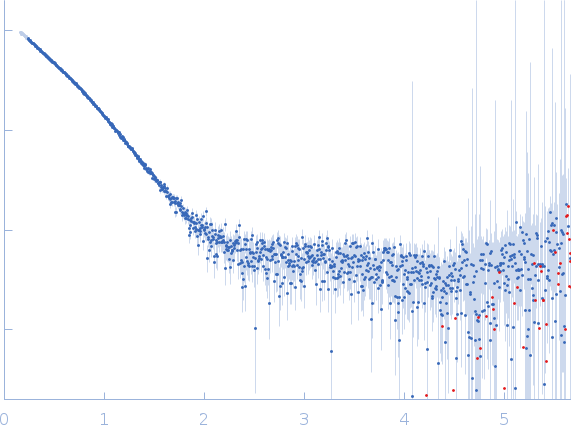
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQR8_fit1_model1.png)
|
| Sample: |
Huntingtin monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20mM BisTris-HCl, 150mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 3
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
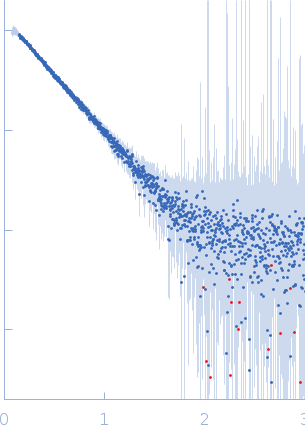
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQS8_fit1_model1.png)
|
| Sample: |
Huntingtin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20mM BisTris-HCl, 150mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 28
|
|
|
|

![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQR8_fit1_model1.png)
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQS8_fit1_model1.png)