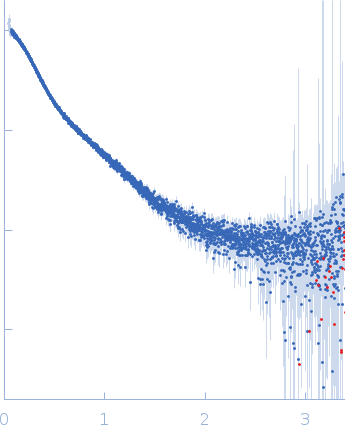
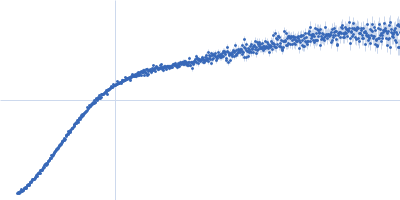
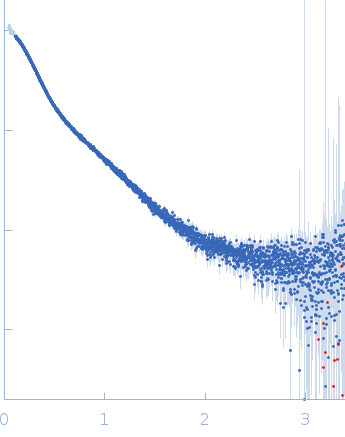
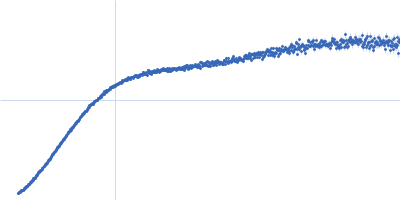
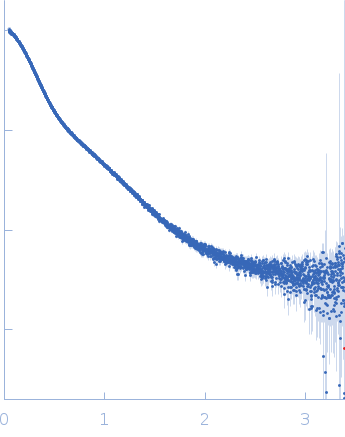
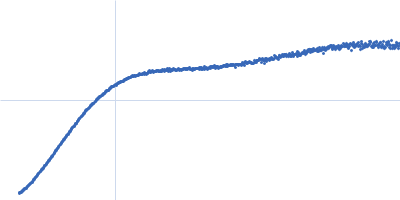
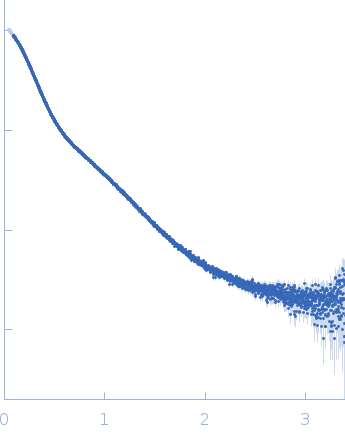
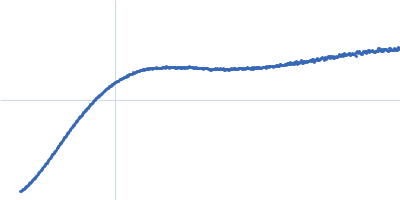
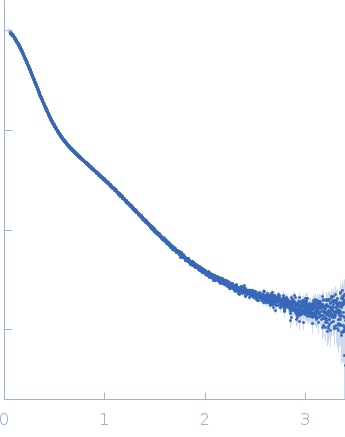
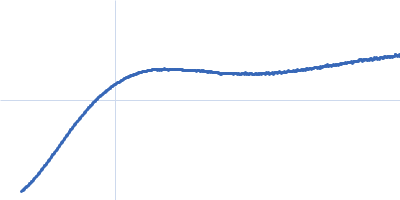
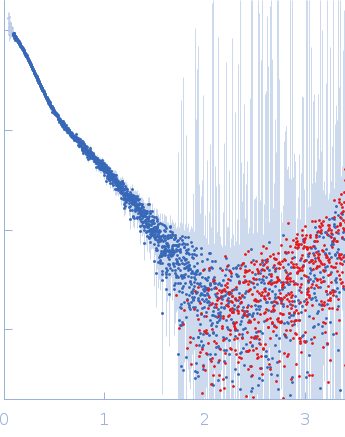
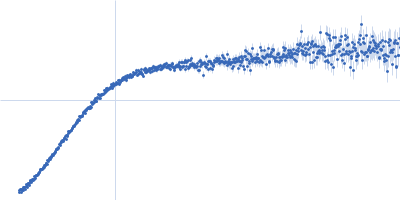
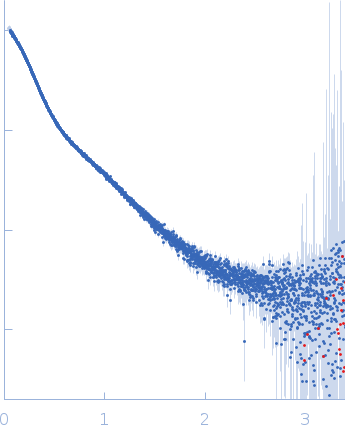
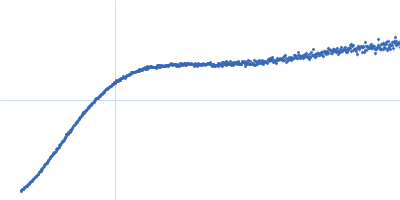
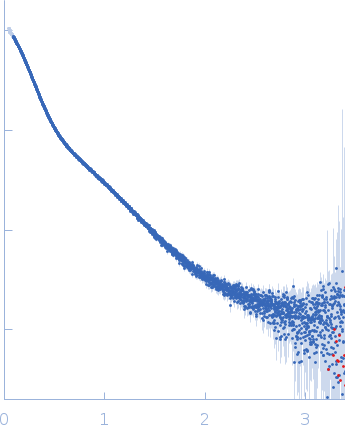
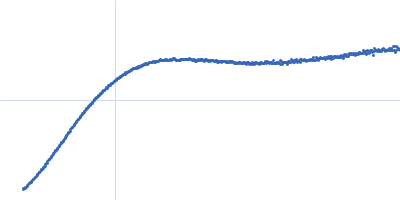
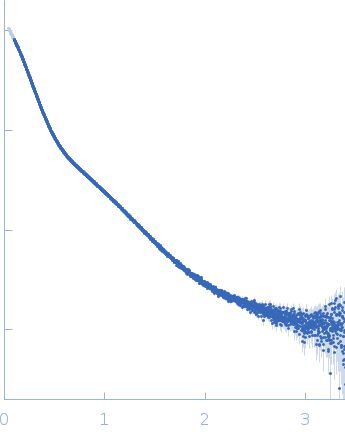
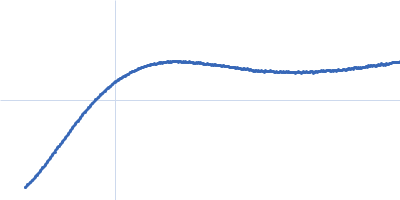
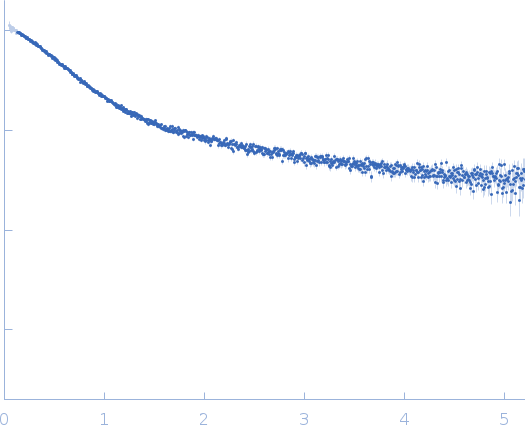
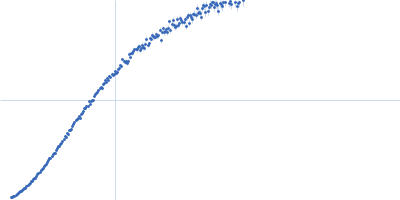
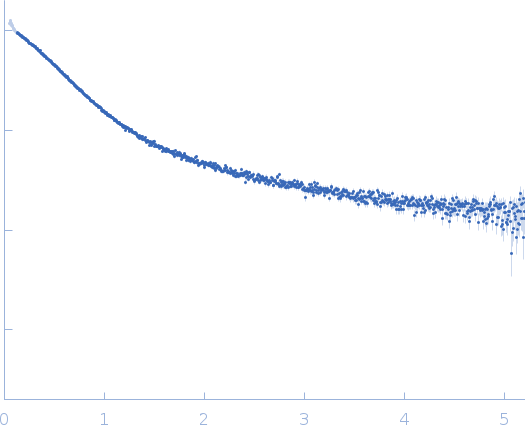
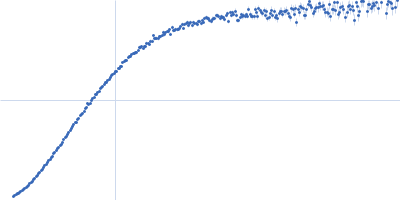
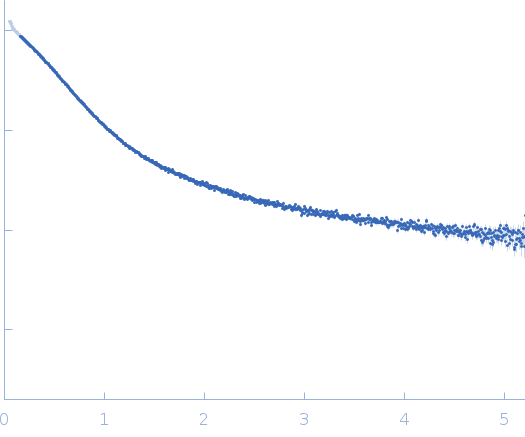
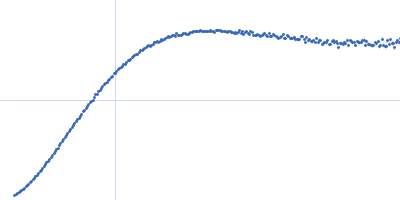
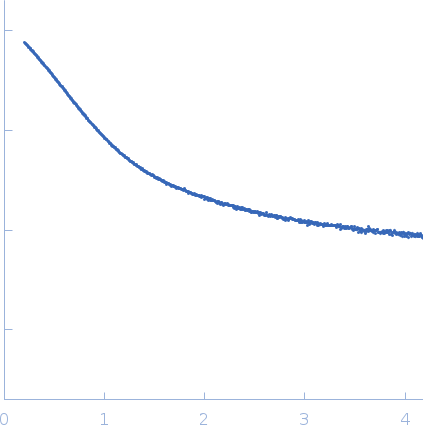
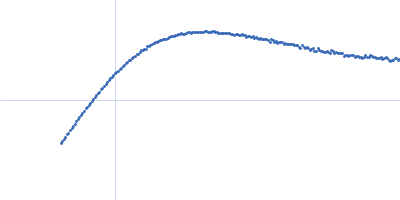
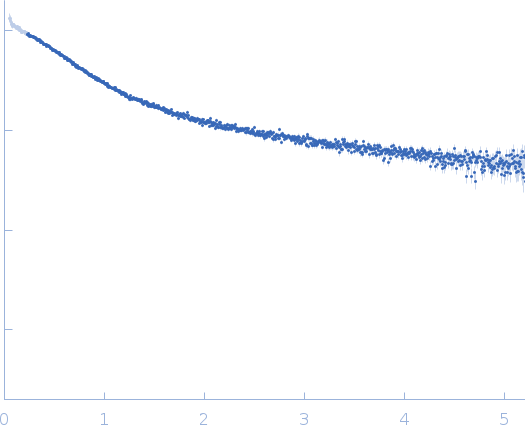
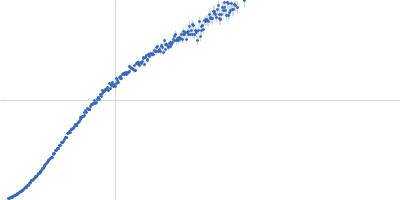
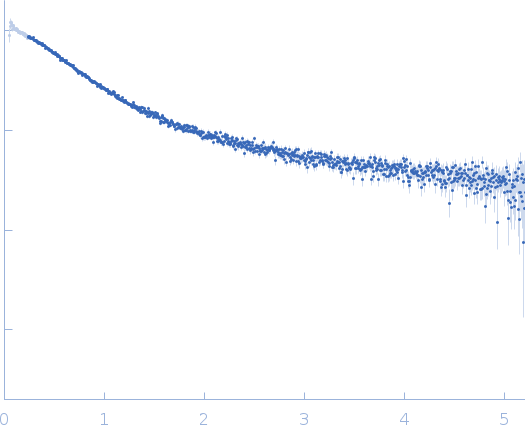
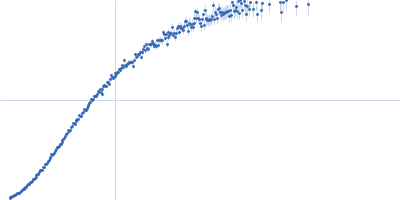
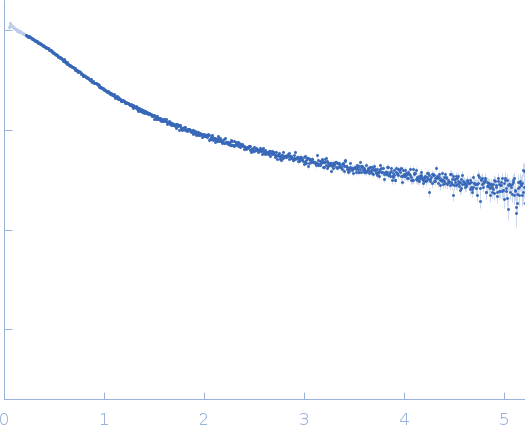
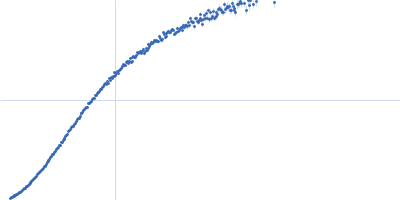
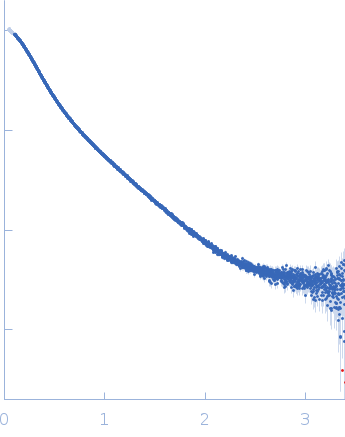
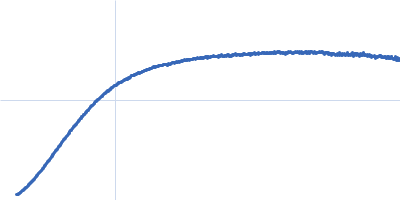
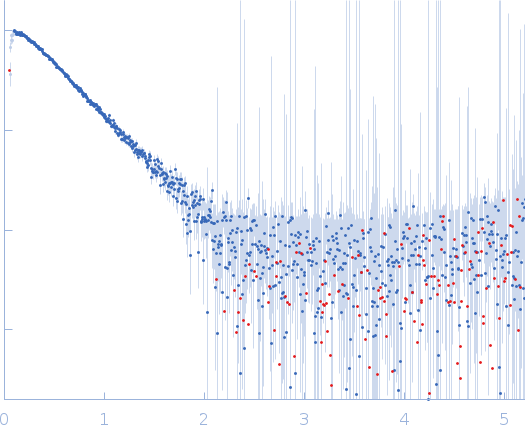
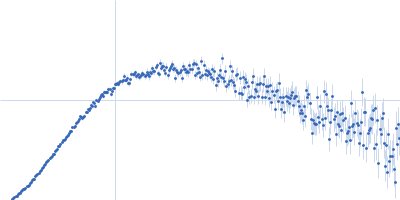
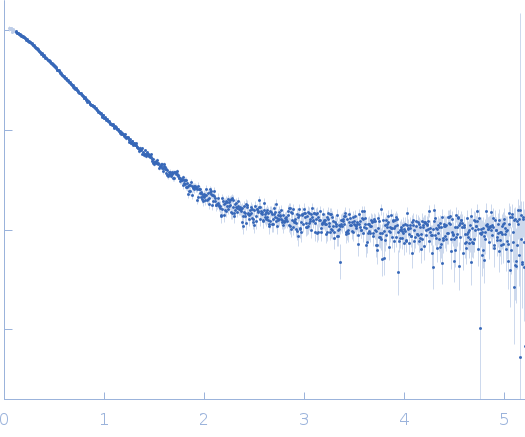
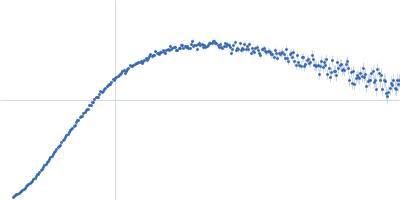
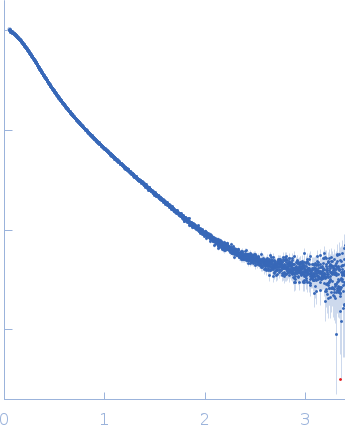
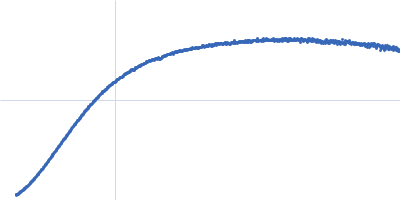
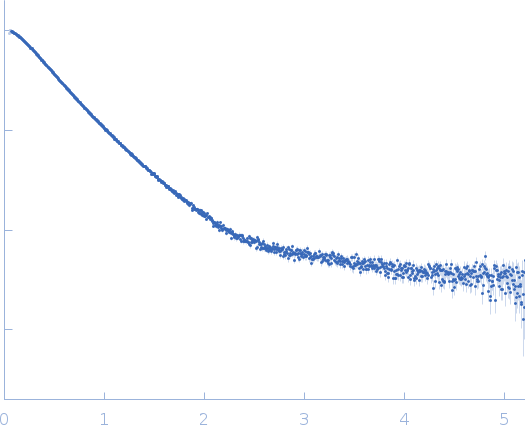
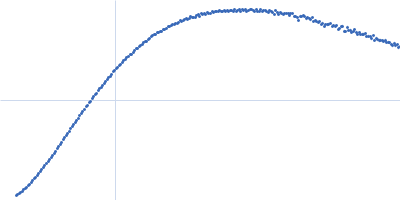
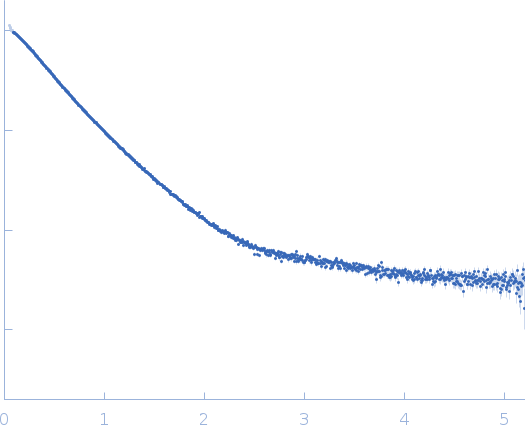
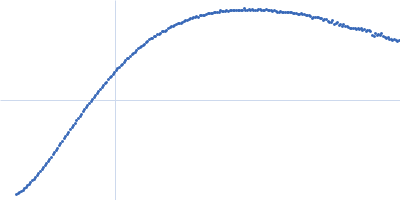
Dynamic ensembles of SARS-CoV-2 N-protein reveal head-to-head coiled-coil-driven oligomerization and phase separation.
Hernandez G,
Martins ML,
Fernandes NP,
Veloso T,
Lopes J,
Gomes T,
Cordeiro TN
Nucleic Acids Res
53(11)
(2025 Jun 6)
|
|
|
|
|
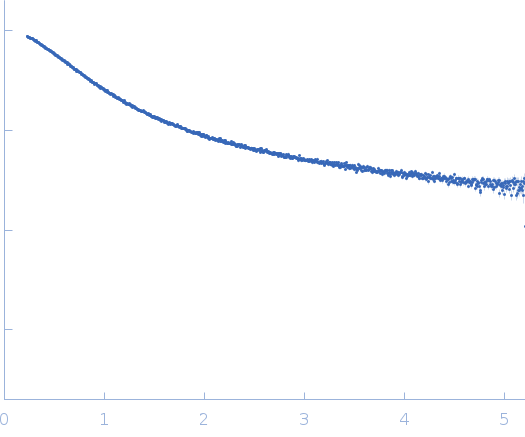
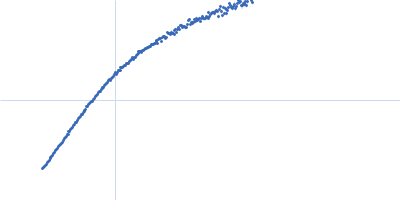
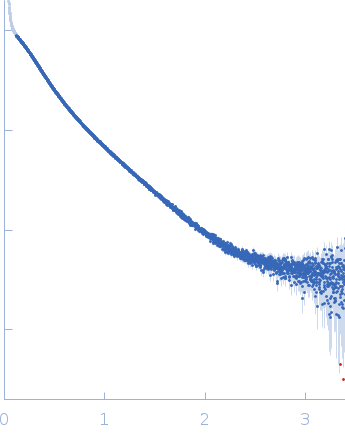
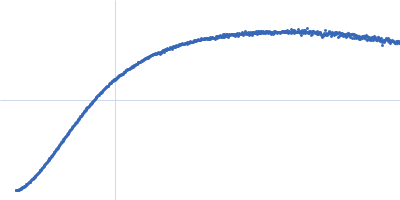
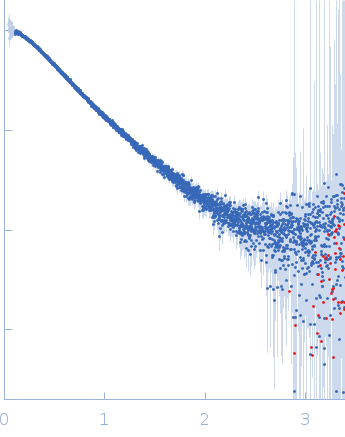
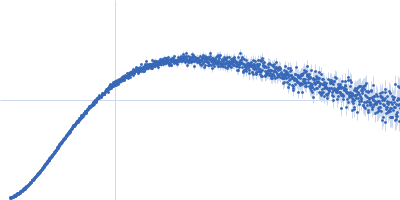
| Sample: |
Nucleoprotein dimer, 82 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
5.7 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
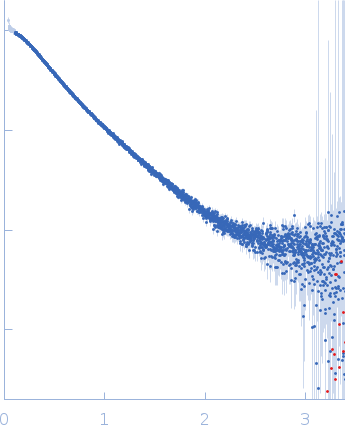
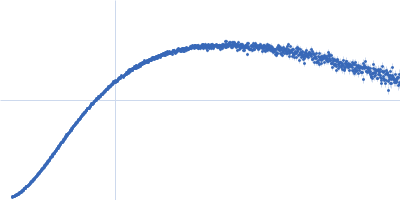
| Sample: |
Nucleoprotein tetramer, 163 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
5.8 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 163 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
6.2 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
215 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 163 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
6.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
578 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein hexamer, 245 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
7.1 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
708 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 187 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
6.3 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 187 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
6.9 |
nm |
| Dmax |
28.5 |
nm |
| VolumePorod |
571 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein hexamer, 281 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
7.6 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
764 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein octamer, 375 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
8.2 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1028 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein dimer, 17 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
4.1 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein dimer, 17 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
4.5 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 8 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jan 29
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 110 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein dimer, 55 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein tetramer, 110 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jan 29
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jan 29
|
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein dimer, 55 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jan 29
|
|
| RgGuinier |
4.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jan 29
|
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
75 |
nm3 |
|
|