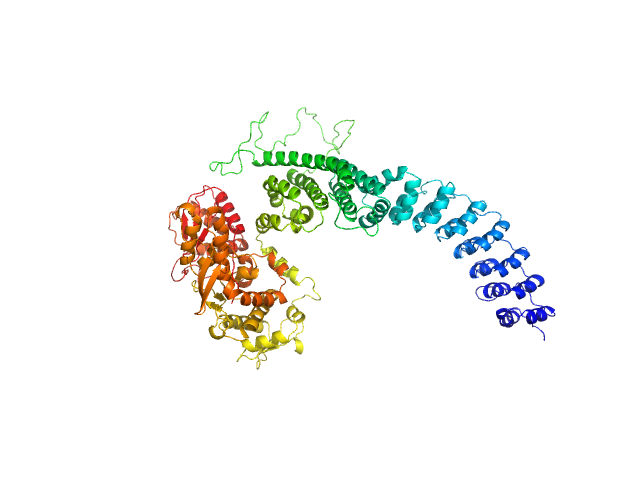
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Düring J,
Wolter M,
Toplak J,
Torres C,
Dybkov O,
Fokkens T,
Bohnsack K,
Urlaub H,
Steinchen W,
Dienemann C,
Lorenz S
Nature Structural & Molecular Biology
(2024 Feb 08)
|
|
|
|

|
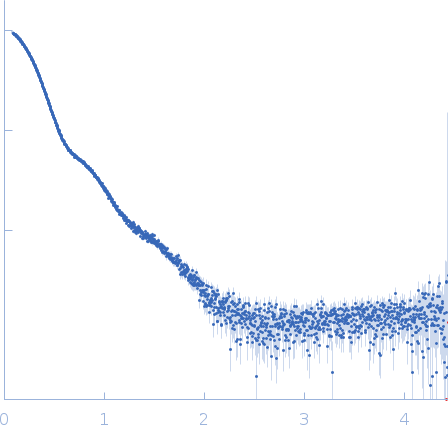
| Sample: |
E3 ubiquitin-protein ligase HACE1 dimer, 205 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
379 |
nm3 |
|
|
|
|
|
|
|
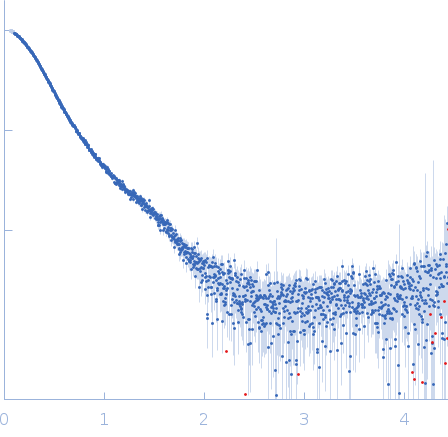
| Sample: |
E3 ubiquitin-protein ligase HACE1 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
219 |
nm3 |
|
|