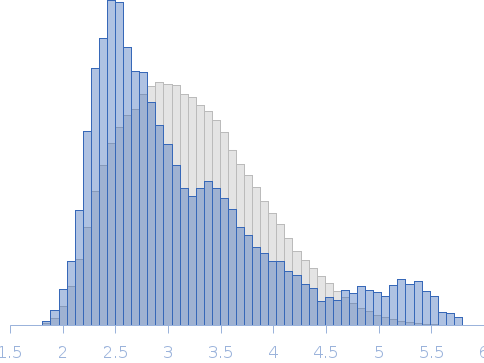
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Sargent R,
Liu DH,
Yadav R,
Glennenmeier D,
Bradford C,
Urbina N,
Beck MR
Protein Sci
34(5):e70122
(2025 May)
|
|
|
|
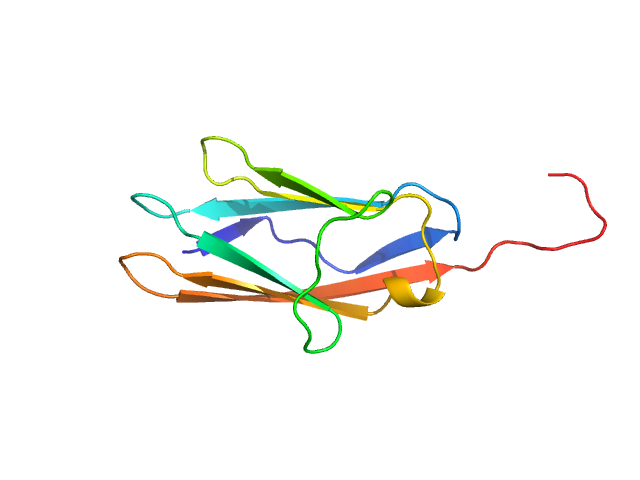
|
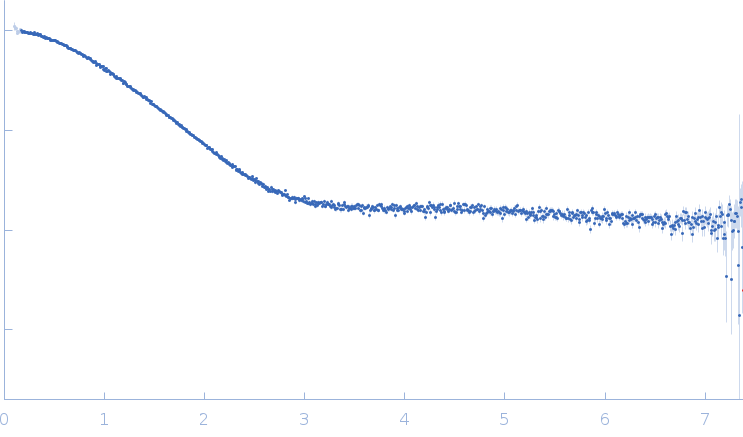
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
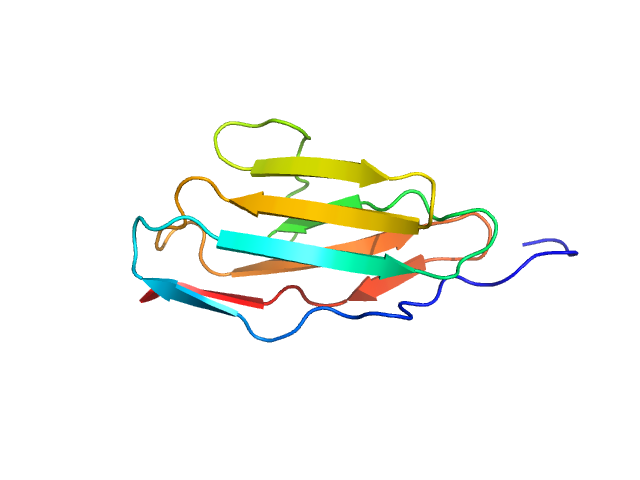
|
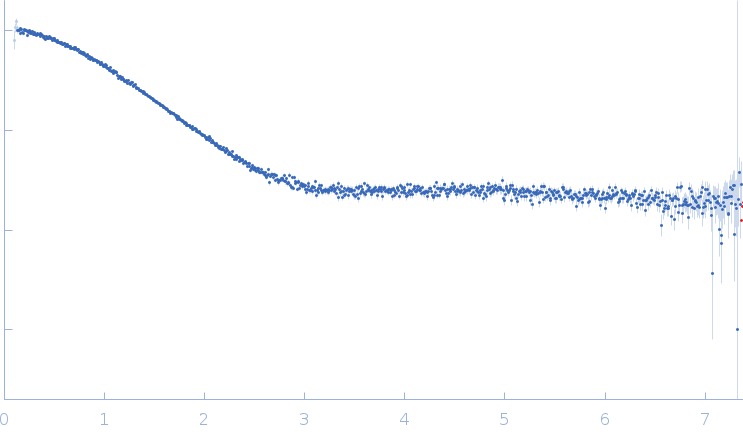
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
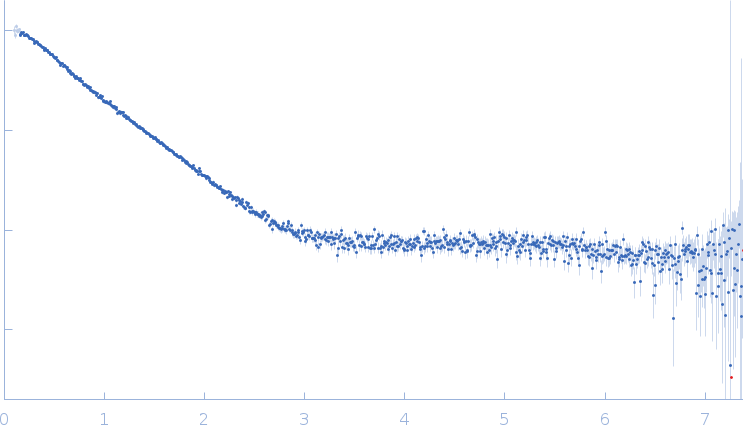
| Sample: |
Palladin monomer, 27 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|