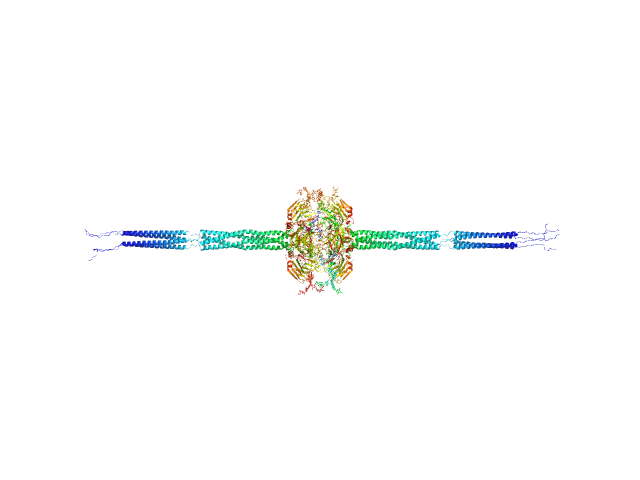
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Shah A,
Zhang X,
Snee M,
Lockhart-Cairns M,
Levy C,
Jowitt T,
Birchenough H,
Dean L,
Collins R,
Dodd R,
Roberts A,
Enghild J,
Mantovani A,
Fontana J,
Baldock C,
Inforzato A,
Richter R,
Day A
Matrix Biology
136:52-68
(2025 Apr)
|
|
|
|

|
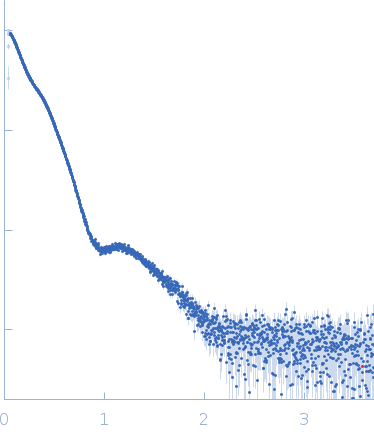
| Sample: |
Pentraxin-related protein PTX3 octamer, 321 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 28
|
|
| RgGuinier |
8.6 |
nm |
| Dmax |
43.1 |
nm |
| VolumePorod |
658 |
nm3 |
|
|
|
|
|
|
|
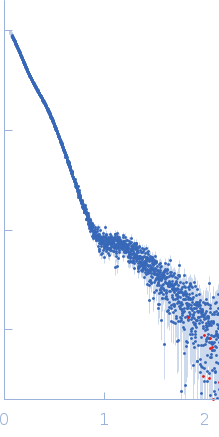
| Sample: |
Pentraxin-related protein PTX3 (N-terminal deletion mutant) octamer, 301 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
|
| RgGuinier |
8.4 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
647 |
nm3 |
|
|
|
|
|
|
|
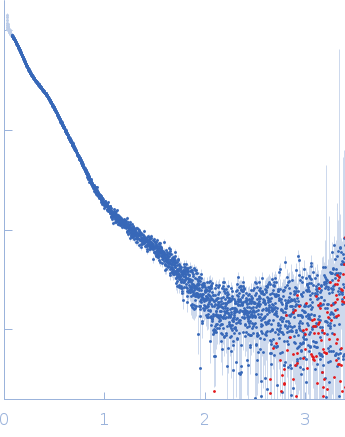
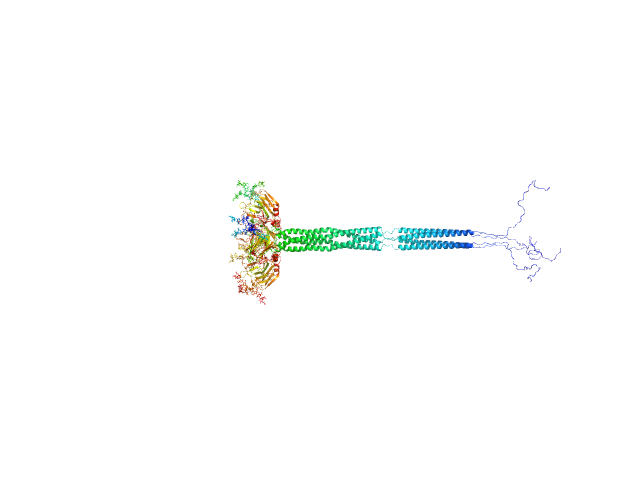
| Sample: |
Pentraxin-related protein PTX3 (C317S, C318S) tetramer, 160 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|