Structural, biophysical and biochemical studies of CAZYmes involved in exopolysaccharides degradation of microbial biofilms
Amanda Freitas Cruz
Universidade de São Paulo
Master's thesis
(2025 Sep 18)
|
|
|
|

|
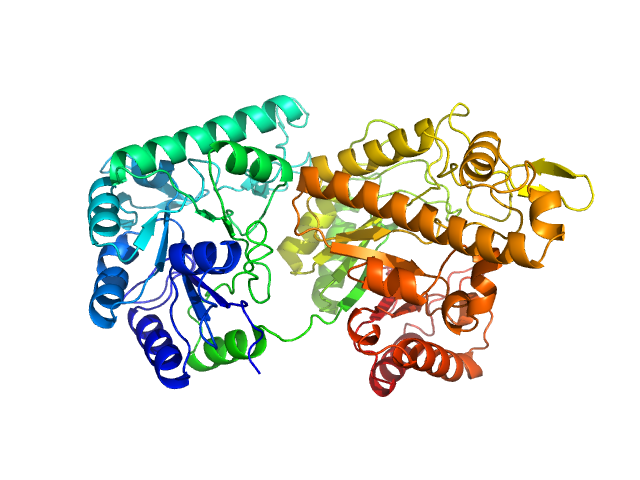
| Sample: |
Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB monomer, 73 kDa Serratia marcescens protein
|
| Buffer: |
phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Feb 3
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB monomer, 69 kDa Serratia marcescens protein
|
| Buffer: |
phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2025 Feb 3
|
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|




