Crystal structure of a complete threonyl tRNA synthetase: toggle clamp model?
Tran T,
Fenwick M,
Edwards T,
Dranow D,
Abendroth J,
Shek R,
Tillery L,
Craig J,
Van Voorhis W,
Myler P
Biochemical and Biophysical Research Communications
:152677
(2025 Sep)
|
|
|
|
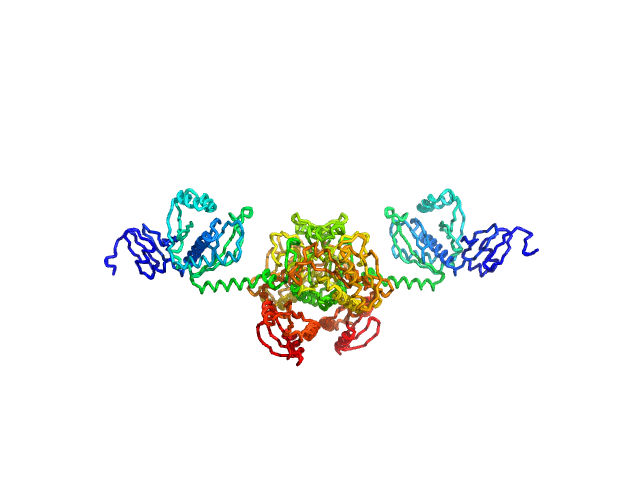
|
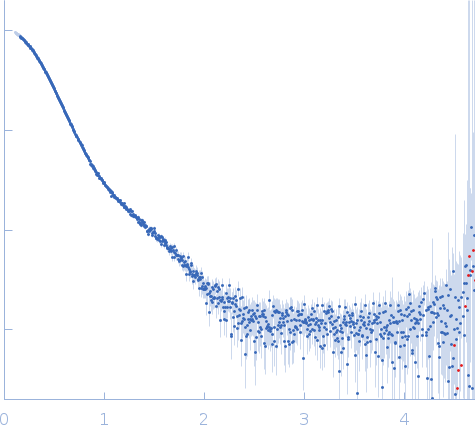
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa Stenotrophomonas maltophilia (strain … protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.2 |
nm |
|
|
|
|
|
|
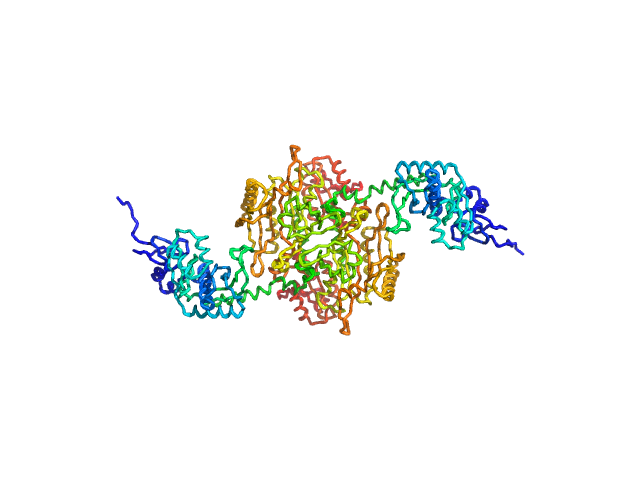
|
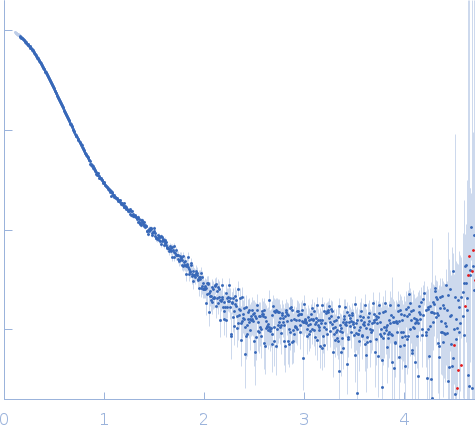
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa Stenotrophomonas maltophilia (strain … protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.2 |
nm |
|
|
|
|
|
|
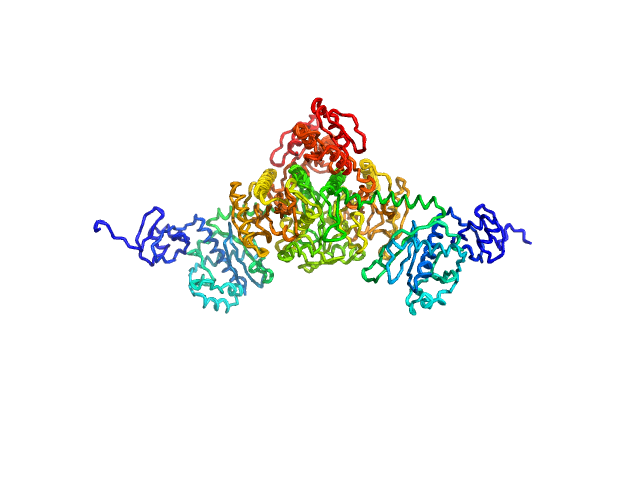
|
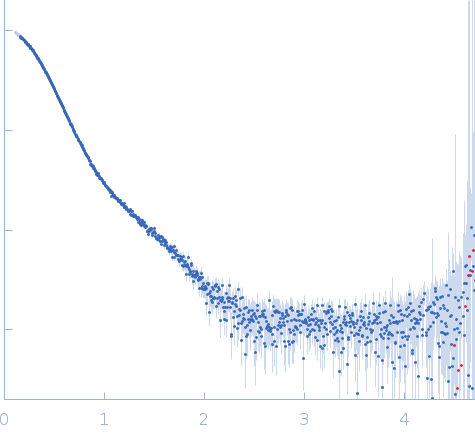
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa Stenotrophomonas maltophilia (strain … protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.2 |
nm |
|
|
|
|
|
|
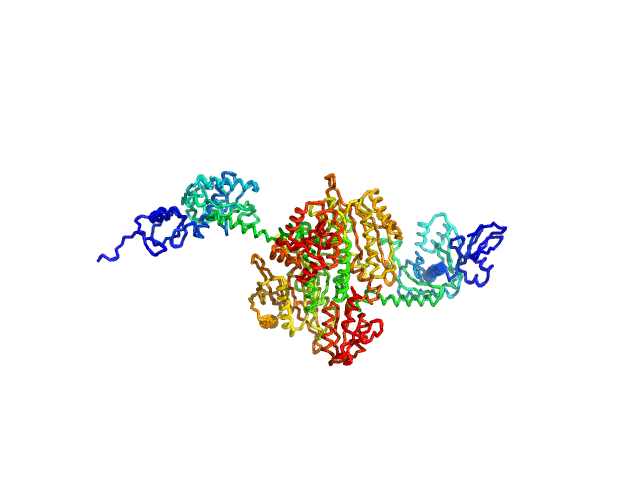
|
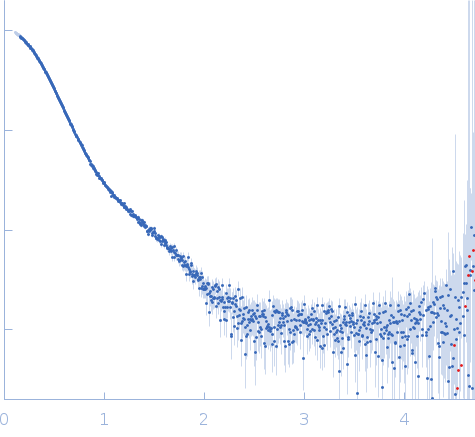
| Sample: |
Stenotrophomonas maltophilia threonyl tRNA synthetase dimer, 146 kDa protein
|
| Buffer: |
20 mM HEPES, 0.2 M NaCl, 1.5% v/v glycerol, and 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2024 Nov 7
|
|
|
|