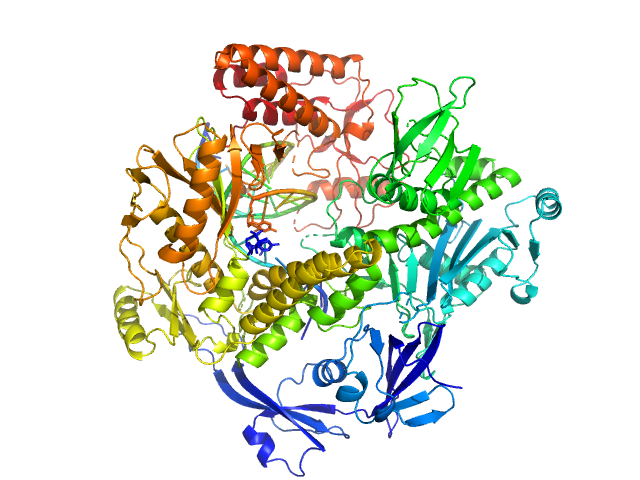
The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Tarbouriech N,
Ducournau C,
Hutin S,
Mas PJ,
Man P,
Forest E,
Hart DJ,
Peyrefitte CN,
Burmeister WP,
Iseni F
Nat Commun
8(1):1455
(2017 Nov 13)
|
|
|
|
|
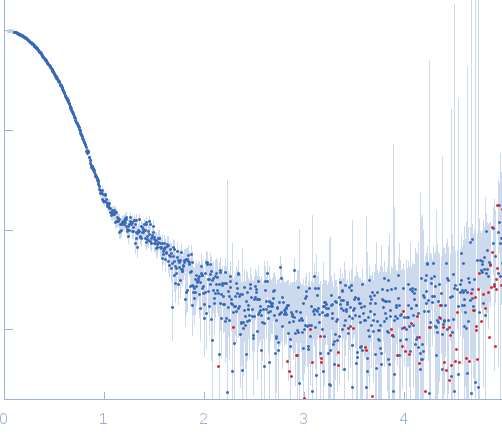
| Sample: |
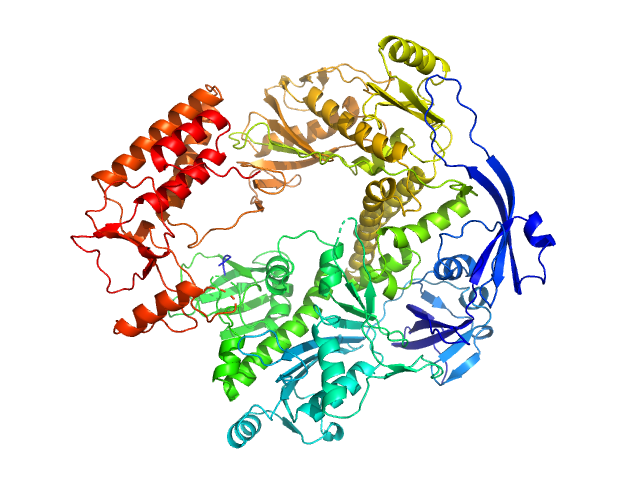
DNA polymerase E9 exo- mutant monomer, 117 kDa Vaccinia virus protein
DNA oligomer template-primer hairpin monomer, 9 kDa synthetic construct DNA
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 12
|
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
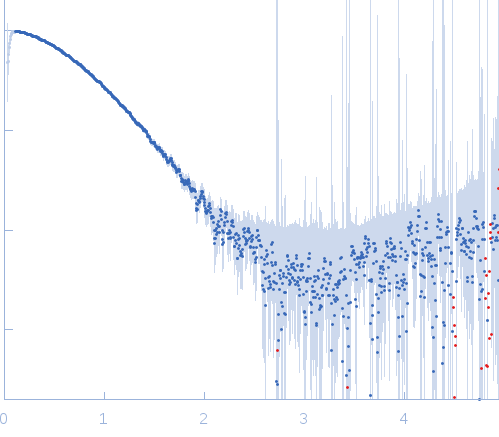
| Sample: |
DNA polymerase E9 monomer, 117 kDa Vaccinia virus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 4 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 27
|
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
|
|
|
|

|
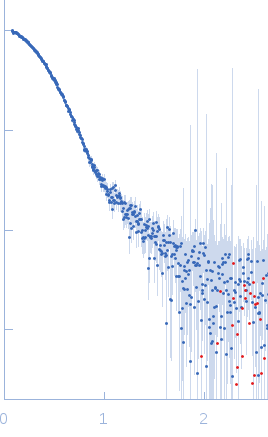
| Sample: |
DNA polymerase processivity factor component A20 C-ter fragment monomer, 17 kDa Vaccinia virus protein
|
| Buffer: |
25 mM Tris-HCl, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
31 |
nm3 |
|
|