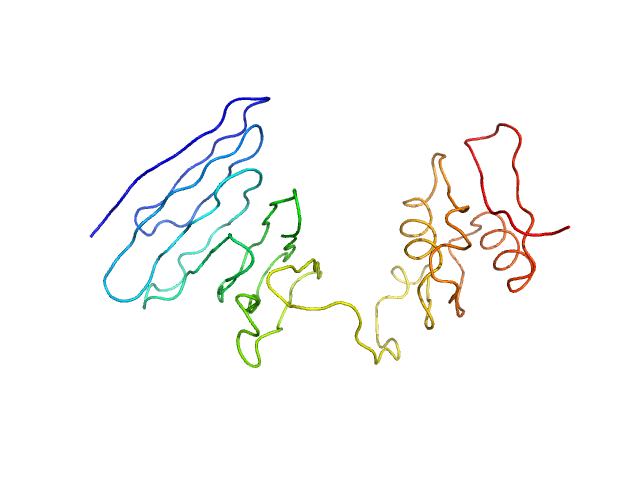
Structural basis of cell surface signaling by a conserved sigma regulator in Gram-negative bacteria.
Jensen JL,
Jernberg BD,
Sinha S,
Colbert CL
J Biol Chem
(2020 Feb 26)
|
|
|
|
|
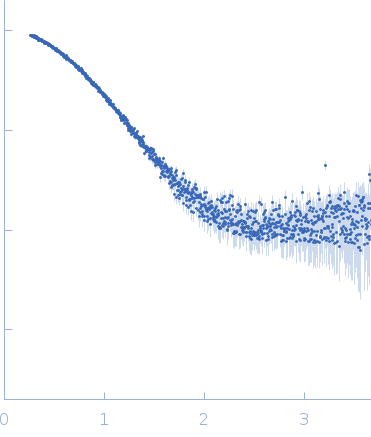
| Sample: |
PupR protein monomer, 24 kDa Pseudomonas putida protein
|
| Buffer: |
25 mM HEPES 400 mM LiCl 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Mar 16
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
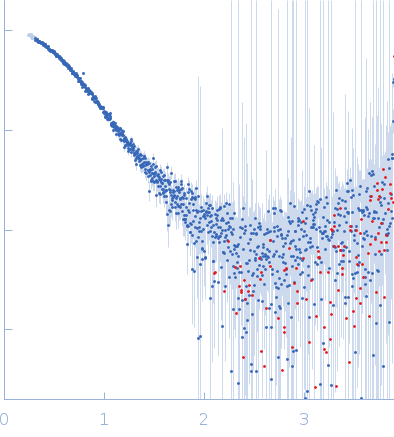
| Sample: |
PupR protein monomer, 24 kDa Pseudomonas putida protein
Ferric-pseudobactin BN7/BN8 receptor monomer, 8 kDa Pseudomonas putida protein
|
| Buffer: |
25 mM HEPES 400 mM LiCl 10% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Mar 16
|
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|