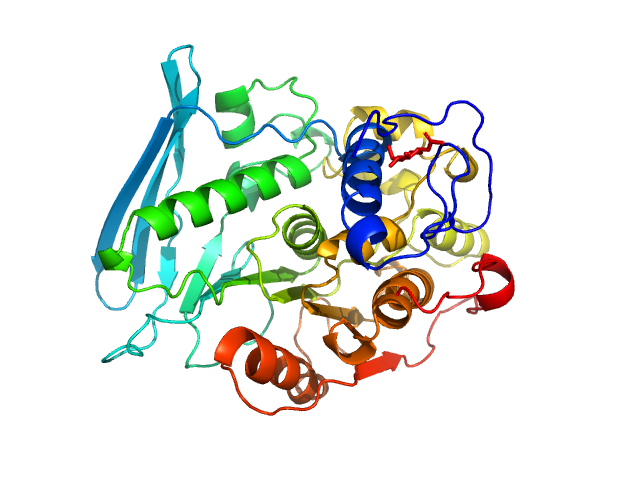
The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Ernst HA,
Mosbech C,
Langkilde AE,
Westh P,
Meyer AS,
Agger JW,
Larsen S
Nat Commun
11(1):1026
(2020 Feb 24)
|
|
|
|
|
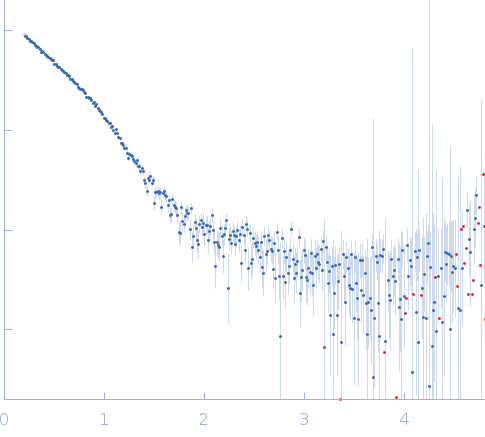
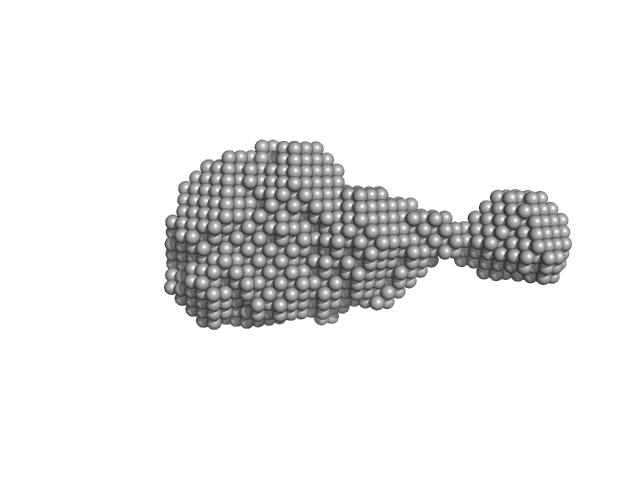
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase) monomer, 51 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
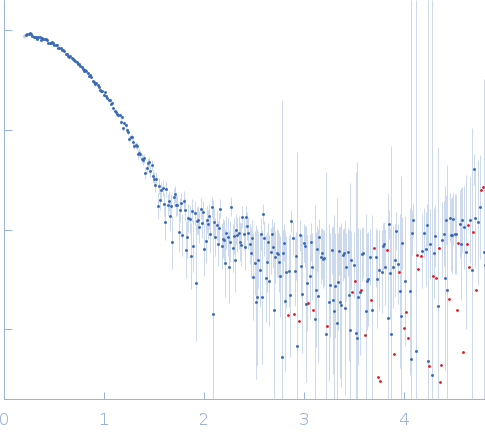
| Sample: |
4-O-methyl-glucuronoyl methylesterase (Glucuronoyl esterase, truncated) monomer, 43 kDa Cerrena unicolor protein
|
| Buffer: |
20 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 10
|
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
50 |
nm3 |
|
|