|
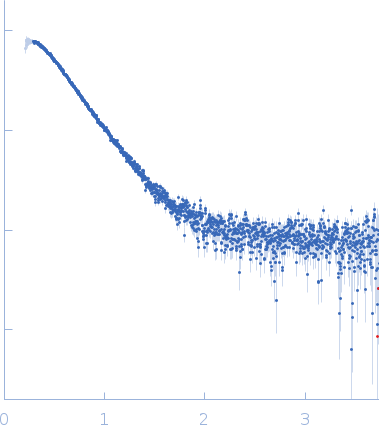
Synchrotron SAXS data from solutions of cold-active AMS8 lipase (without polyhistidine affinity tag) in 50 mM Tris HCl, 5 mM CaCl2, pH 8 were collected on the BL1.3W SAXS/WAXS beam line at the Synchrotron Light Research Institute (SLRI; Nakhon Ratchasima, Thailand) using a MAR 345 Image Plate detector at a sample-detector distance of 1.7 m and at a wavelength of λ = 0.138 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 2.00 mg/ml was measured at 25°C. One 600 second frame was collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
|
|
LipAMS8
(AMS8)
|
| Mol. type |
|
Protein |
| Organism |
|
Pseudomonas sp. AMS8 |
| Olig. state |
|
Monomer |
| Mon. MW |
|
50.0 kDa |
| |
| UniProt |
|
E1B2U7
(1-476)
|
| Sequence |
|
FASTA |
| |
|
 s, nm-1
s, nm-1